> Biological Physics | Proteins | Comp Bio | ML
https://scholar.google.com/citations?user=n55NtEsAAAAJ&hl=en



colab.research.google.com/github/eagal...

colab.research.google.com/github/eagal...
6/n

6/n
5/n
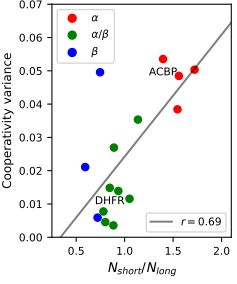
5/n
4/n

4/n
3/n

3/n
2/n
2/n

