| PhD from PlevkaLab | #StructuralBiology of #Viruses #HIV | #CryoEM | #Memes
Twitter/X: @F4ustus
m.xkcd.com/3056/

m.xkcd.com/3056/
m.xkcd.com/3056/

m.xkcd.com/3056/




Read more in Nature: www.nature.com/articles/s41... 🧪

Read more in Nature: www.nature.com/articles/s41... 🧪







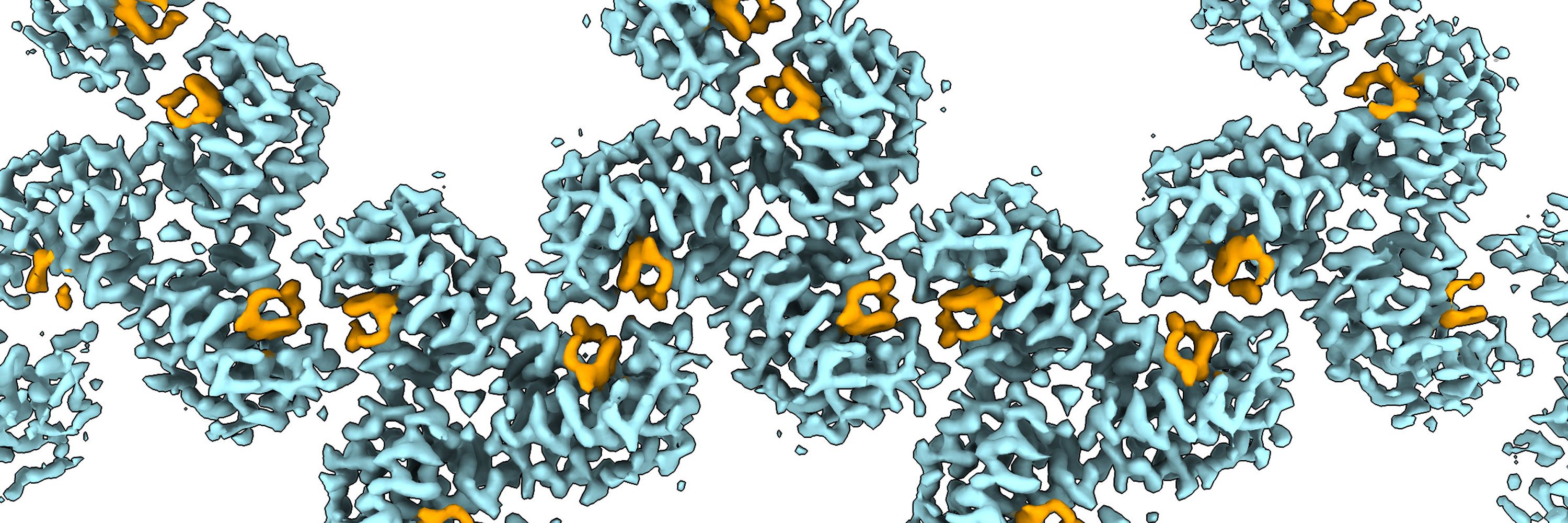
In the meantime, we analyzed MA from “HIV-1 Gag cleavage mutant” virions. One can block the protease recognition site in Gag by mutation. We speculated that one of the cleavage sites is an MA maturation trigger. Surprisingly, the common denominator for the mature matrix was a free SP2 peptide! 8/x

In the meantime, we analyzed MA from “HIV-1 Gag cleavage mutant” virions. One can block the protease recognition site in Gag by mutation. We speculated that one of the cleavage sites is an MA maturation trigger. Surprisingly, the common denominator for the mature matrix was a free SP2 peptide! 8/x

We trained a crYOLO model that can recognise the features of an HIV-1 virion and oversample the virus surface. Then, we used the known matrix structure from tomography to initialise the reconstruction and classification. 6/x
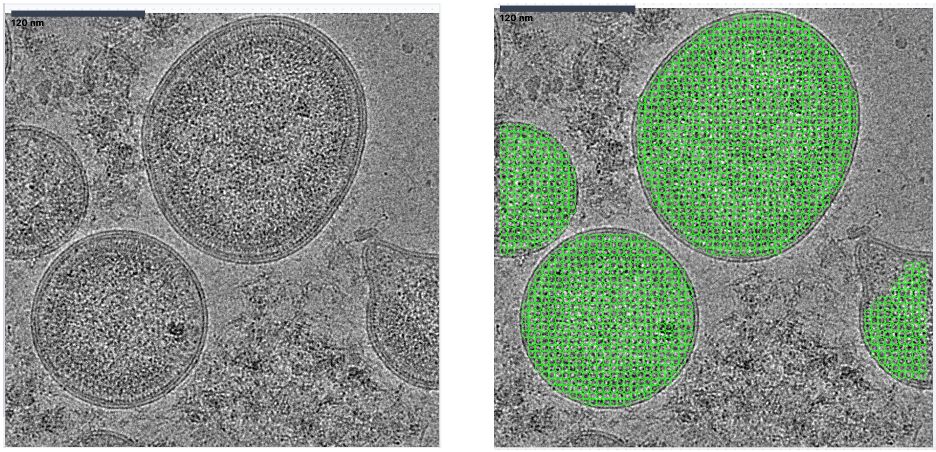
We trained a crYOLO model that can recognise the features of an HIV-1 virion and oversample the virus surface. Then, we used the known matrix structure from tomography to initialise the reconstruction and classification. 6/x


Paper: tinyurl.com/c7szhtfa
Paper: tinyurl.com/c7szhtfa





