Daniel Hurdiss
@danielhurdiss.bsky.social
Assistant Professor at Utrecht University 🇳🇱
We use #cryoEM and molecular #virology techniques to study the infection mechanisms of +ssRNA viruses.
https://www.uu.nl/medewerkers/DLHurdiss
Views are my own.
We use #cryoEM and molecular #virology techniques to study the infection mechanisms of +ssRNA viruses.
https://www.uu.nl/medewerkers/DLHurdiss
Views are my own.
Rounding off an incredibly busy week with the excellent third installment of the Dutch Structural Biology Meeting! Many thanks to Arjen for the kind invitation to present our lab’s work.

September 26, 2025 at 6:37 PM
Rounding off an incredibly busy week with the excellent third installment of the Dutch Structural Biology Meeting! Many thanks to Arjen for the kind invitation to present our lab’s work.
Back from the 9th International Calicivirus Conference in Banff! Robin Veenstra and I really enjoyed both the stunning mountain views and the exciting science being shared in the field.
Thanks to the organisers for putting together this great meeting. Lots of exciting collaborations ahead!
Thanks to the organisers for putting together this great meeting. Lots of exciting collaborations ahead!


September 13, 2025 at 9:09 AM
Back from the 9th International Calicivirus Conference in Banff! Robin Veenstra and I really enjoyed both the stunning mountain views and the exciting science being shared in the field.
Thanks to the organisers for putting together this great meeting. Lots of exciting collaborations ahead!
Thanks to the organisers for putting together this great meeting. Lots of exciting collaborations ahead!
Our model could help reconcile noroviruses, and perhaps enteroviruses, with other positive-strand RNA viruses that use large “crown” complexes to couple genome replication with RNA export.
NS3 may represent a minimalist alternative to this strategy.
NS3 may represent a minimalist alternative to this strategy.

June 18, 2025 at 12:23 PM
Our model could help reconcile noroviruses, and perhaps enteroviruses, with other positive-strand RNA viruses that use large “crown” complexes to couple genome replication with RNA export.
NS3 may represent a minimalist alternative to this strategy.
NS3 may represent a minimalist alternative to this strategy.
Putting all of this together, we propose that NS3 transports newly synthesised plus-strand RNA from within single-membrane vesicles.
Once exported, the RNA can be translated or packaged in the cytoplasm.
A streamlined mechanism for linking replication to the next steps.
Once exported, the RNA can be translated or packaged in the cytoplasm.
A streamlined mechanism for linking replication to the next steps.

June 18, 2025 at 12:12 PM
Putting all of this together, we propose that NS3 transports newly synthesised plus-strand RNA from within single-membrane vesicles.
Once exported, the RNA can be translated or packaged in the cytoplasm.
A streamlined mechanism for linking replication to the next steps.
Once exported, the RNA can be translated or packaged in the cytoplasm.
A streamlined mechanism for linking replication to the next steps.
Similar predictions of NS3 and its enterovirus homolog 2C, including a 50-mer polyA strand, suggest a plausible path for RNA translocation.
This may point to a conserved mechanism among SF3 ATPases in positive-strand RNA viruses (but further validation is needed)!
This may point to a conserved mechanism among SF3 ATPases in positive-strand RNA viruses (but further validation is needed)!

June 18, 2025 at 12:07 PM
Similar predictions of NS3 and its enterovirus homolog 2C, including a 50-mer polyA strand, suggest a plausible path for RNA translocation.
This may point to a conserved mechanism among SF3 ATPases in positive-strand RNA viruses (but further validation is needed)!
This may point to a conserved mechanism among SF3 ATPases in positive-strand RNA viruses (but further validation is needed)!
Here is the NTD prediction in all its glory!
A compact, high-confidence transmembrane channel with convincing architecture and hydrophobicity profile.
The pore is positively charged, suggesting it could help guide negatively charged RNA through the membrane.
Looks solid to us!
A compact, high-confidence transmembrane channel with convincing architecture and hydrophobicity profile.
The pore is positively charged, suggesting it could help guide negatively charged RNA through the membrane.
Looks solid to us!

June 18, 2025 at 11:56 AM
Here is the NTD prediction in all its glory!
A compact, high-confidence transmembrane channel with convincing architecture and hydrophobicity profile.
The pore is positively charged, suggesting it could help guide negatively charged RNA through the membrane.
Looks solid to us!
A compact, high-confidence transmembrane channel with convincing architecture and hydrophobicity profile.
The pore is positively charged, suggesting it could help guide negatively charged RNA through the membrane.
Looks solid to us!
Amazingly, this gave us a high-confidence transmembrane channel for both mouse and human NS3, with oleic acids forming a bilayer-like pattern.
Our group now call this the “fatty acid hack.”
The #alphafold predictions with and without fatty acids shown below 👇
Our group now call this the “fatty acid hack.”
The #alphafold predictions with and without fatty acids shown below 👇

June 18, 2025 at 11:47 AM
Amazingly, this gave us a high-confidence transmembrane channel for both mouse and human NS3, with oleic acids forming a bilayer-like pattern.
Our group now call this the “fatty acid hack.”
The #alphafold predictions with and without fatty acids shown below 👇
Our group now call this the “fatty acid hack.”
The #alphafold predictions with and without fatty acids shown below 👇
At this point, the missing piece of the puzzle was the N-terminal membrane-binding domain, which we had replaced with a soluble hexamerization domain to enable structural studies 🧩
To understand the full picture, we turned to #AlphaFold3.
To understand the full picture, we turned to #AlphaFold3.

June 18, 2025 at 11:39 AM
At this point, the missing piece of the puzzle was the N-terminal membrane-binding domain, which we had replaced with a soluble hexamerization domain to enable structural studies 🧩
To understand the full picture, we turned to #AlphaFold3.
To understand the full picture, we turned to #AlphaFold3.
Does our hex-NS3 construct bind RNA? 🧬
Yes!
Single-molecule magnetic tweezers experiments with the @dulinlab.bsky.social show binding to ssRNA and dsRNA, with compaction of the latter.
No ATP-driven helicase activity detected (thoughts on that in the paper).
Yes!
Single-molecule magnetic tweezers experiments with the @dulinlab.bsky.social show binding to ssRNA and dsRNA, with compaction of the latter.
No ATP-driven helicase activity detected (thoughts on that in the paper).

June 18, 2025 at 11:32 AM
Does our hex-NS3 construct bind RNA? 🧬
Yes!
Single-molecule magnetic tweezers experiments with the @dulinlab.bsky.social show binding to ssRNA and dsRNA, with compaction of the latter.
No ATP-driven helicase activity detected (thoughts on that in the paper).
Yes!
Single-molecule magnetic tweezers experiments with the @dulinlab.bsky.social show binding to ssRNA and dsRNA, with compaction of the latter.
No ATP-driven helicase activity detected (thoughts on that in the paper).
The nucleotide-binding sites were particularly well resolved. Clear density for ATPγS, Mg²⁺, coordinating residues, and water molecules! 🤩
A great starting point for structure-based drug design! 💊
A great starting point for structure-based drug design! 💊

June 18, 2025 at 10:10 AM
The nucleotide-binding sites were particularly well resolved. Clear density for ATPγS, Mg²⁺, coordinating residues, and water molecules! 🤩
A great starting point for structure-based drug design! 💊
A great starting point for structure-based drug design! 💊
The NS3 hexamer adopts a split lock-washer conformation, characteristic of AAA+ ATPases that use a hand-over-hand translocation mechanism.
Also, it kind of looks like a Rolo...
Also, it kind of looks like a Rolo...

June 18, 2025 at 10:03 AM
The NS3 hexamer adopts a split lock-washer conformation, characteristic of AAA+ ATPases that use a hand-over-hand translocation mechanism.
Also, it kind of looks like a Rolo...
Also, it kind of looks like a Rolo...
From ~1000 images collected on a Glacios (thank you
@ievadr.bsky.social!), we obtained a 2.9 Å resolution #cryoEM map of NS3 in complex with ATPγS.
This gave us our first high-res look at the norovirus NS3 hexamer in action!
It is a fun (and small) data set. We have uploaded this to EMPIAR.
@ievadr.bsky.social!), we obtained a 2.9 Å resolution #cryoEM map of NS3 in complex with ATPγS.
This gave us our first high-res look at the norovirus NS3 hexamer in action!
It is a fun (and small) data set. We have uploaded this to EMPIAR.

June 18, 2025 at 9:29 AM
From ~1000 images collected on a Glacios (thank you
@ievadr.bsky.social!), we obtained a 2.9 Å resolution #cryoEM map of NS3 in complex with ATPγS.
This gave us our first high-res look at the norovirus NS3 hexamer in action!
It is a fun (and small) data set. We have uploaded this to EMPIAR.
@ievadr.bsky.social!), we obtained a 2.9 Å resolution #cryoEM map of NS3 in complex with ATPγS.
This gave us our first high-res look at the norovirus NS3 hexamer in action!
It is a fun (and small) data set. We have uploaded this to EMPIAR.
Their N-terminal membrane-binding region, essential for proper assembly, has also made them difficult to study in an oligomeric form.
To overcome this, we engineered a soluble, hexameric version of norovirus NS3.
Big thanks to @joostsnijder.bsky.social lab for mass spec/photometry work!
To overcome this, we engineered a soluble, hexameric version of norovirus NS3.
Big thanks to @joostsnijder.bsky.social lab for mass spec/photometry work!

June 18, 2025 at 9:07 AM
Their N-terminal membrane-binding region, essential for proper assembly, has also made them difficult to study in an oligomeric form.
To overcome this, we engineered a soluble, hexameric version of norovirus NS3.
Big thanks to @joostsnijder.bsky.social lab for mass spec/photometry work!
To overcome this, we engineered a soluble, hexameric version of norovirus NS3.
Big thanks to @joostsnijder.bsky.social lab for mass spec/photometry work!
For DNA viruses, pioneering studies have shown how SF3 proteins work as replicative helicases.
They unwind DNA using coordinated, ATP-driven conformational changes across a hexameric ring.
But no equivalent structures are available for SF3 proteins encoded by RNA viruses...
They unwind DNA using coordinated, ATP-driven conformational changes across a hexameric ring.
But no equivalent structures are available for SF3 proteins encoded by RNA viruses...
June 18, 2025 at 8:24 AM
For DNA viruses, pioneering studies have shown how SF3 proteins work as replicative helicases.
They unwind DNA using coordinated, ATP-driven conformational changes across a hexameric ring.
But no equivalent structures are available for SF3 proteins encoded by RNA viruses...
They unwind DNA using coordinated, ATP-driven conformational changes across a hexameric ring.
But no equivalent structures are available for SF3 proteins encoded by RNA viruses...
Latest preprint from the lab, many years in the making!
By combining #cryoEM with #AlphaFold3 modelling, we propose that norovirus NS3 forms a transmembrane RNA translocase.
This could have big implications for our understanding of viral replication & assembly (🧵)
www.biorxiv.org/content/10.1...
By combining #cryoEM with #AlphaFold3 modelling, we propose that norovirus NS3 forms a transmembrane RNA translocase.
This could have big implications for our understanding of viral replication & assembly (🧵)
www.biorxiv.org/content/10.1...
June 18, 2025 at 6:32 AM
Latest preprint from the lab, many years in the making!
By combining #cryoEM with #AlphaFold3 modelling, we propose that norovirus NS3 forms a transmembrane RNA translocase.
This could have big implications for our understanding of viral replication & assembly (🧵)
www.biorxiv.org/content/10.1...
By combining #cryoEM with #AlphaFold3 modelling, we propose that norovirus NS3 forms a transmembrane RNA translocase.
This could have big implications for our understanding of viral replication & assembly (🧵)
www.biorxiv.org/content/10.1...
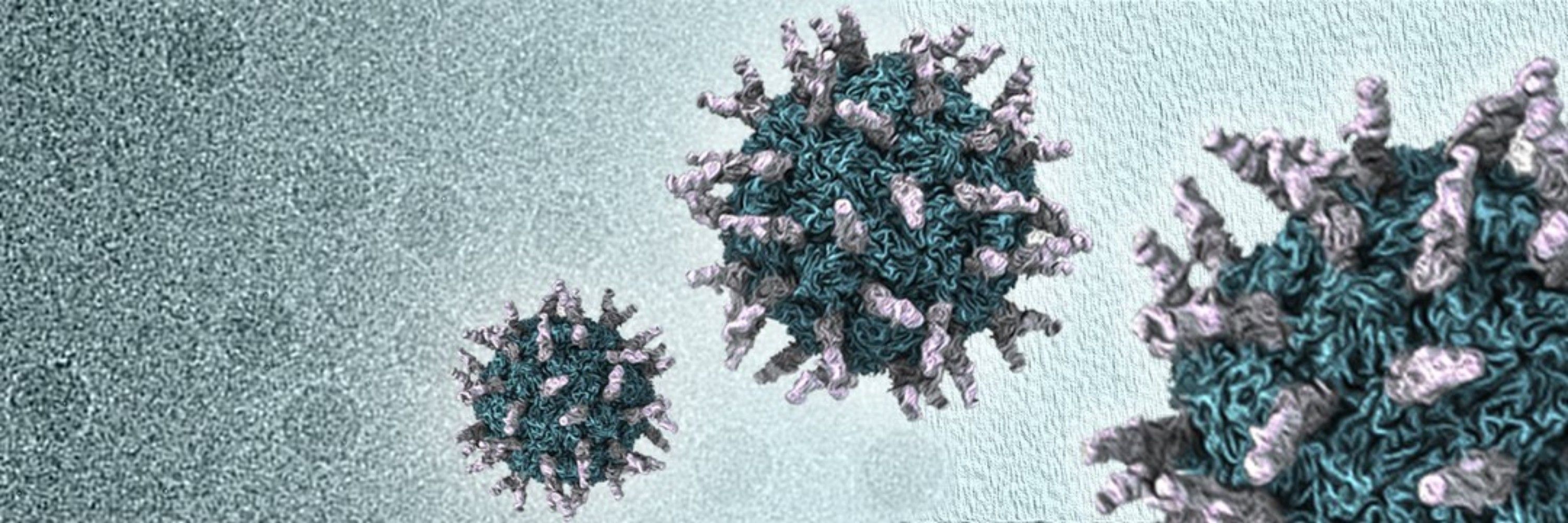
Think coronavirus spikes have run out of surprises? Think again.
Our latest preprint dives into the highly unusual spikes of marine mammal coronaviruses.
www.biorxiv.org/content/10.1...
This #cryoEM study was led by @viralfusion.bsky.social, with key contributions from an amazing team.
Our latest preprint dives into the highly unusual spikes of marine mammal coronaviruses.
www.biorxiv.org/content/10.1...
This #cryoEM study was led by @viralfusion.bsky.social, with key contributions from an amazing team.
May 23, 2025 at 5:02 AM
Think coronavirus spikes have run out of surprises? Think again.
Our latest preprint dives into the highly unusual spikes of marine mammal coronaviruses.
www.biorxiv.org/content/10.1...
This #cryoEM study was led by @viralfusion.bsky.social, with key contributions from an amazing team.
Our latest preprint dives into the highly unusual spikes of marine mammal coronaviruses.
www.biorxiv.org/content/10.1...
This #cryoEM study was led by @viralfusion.bsky.social, with key contributions from an amazing team.
Very proud of Judith Ahr who gave her final MSc internship presentation this week!
Judith led an AI-driven antiviral biologics project with incredible independence. In addition, she supported many others in the lab with her expertise.
We’ll miss you and can’t wait to see what’s next!
Judith led an AI-driven antiviral biologics project with incredible independence. In addition, she supported many others in the lab with her expertise.
We’ll miss you and can’t wait to see what’s next!

May 21, 2025 at 8:18 AM
Very proud of Judith Ahr who gave her final MSc internship presentation this week!
Judith led an AI-driven antiviral biologics project with incredible independence. In addition, she supported many others in the lab with her expertise.
We’ll miss you and can’t wait to see what’s next!
Judith led an AI-driven antiviral biologics project with incredible independence. In addition, she supported many others in the lab with her expertise.
We’ll miss you and can’t wait to see what’s next!
Height and youth proved too much for me on this occasion, but we all put in strong sub-50 minute times! 😃

May 19, 2025 at 6:15 PM
Height and youth proved too much for me on this occasion, but we all put in strong sub-50 minute times! 😃
I’m delighted that we could contribute the #cryoEM data to this incredible antiviral discovery project from the group of @neytsjohan.bsky.social 💊
"A coronavirus assembly inhibitor that targets the viral membrane protein"
www.nature.com/articles/s41...
A huge congrats to everyone involved! 🎉
"A coronavirus assembly inhibitor that targets the viral membrane protein"
www.nature.com/articles/s41...
A huge congrats to everyone involved! 🎉
March 26, 2025 at 4:31 PM
I’m delighted that we could contribute the #cryoEM data to this incredible antiviral discovery project from the group of @neytsjohan.bsky.social 💊
"A coronavirus assembly inhibitor that targets the viral membrane protein"
www.nature.com/articles/s41...
A huge congrats to everyone involved! 🎉
"A coronavirus assembly inhibitor that targets the viral membrane protein"
www.nature.com/articles/s41...
A huge congrats to everyone involved! 🎉
Our third GPU workstation is up and running!
Excited to see what this can churn out in the next few years 🚀
Excited to see what this can churn out in the next few years 🚀

March 13, 2025 at 10:54 AM
Our third GPU workstation is up and running!
Excited to see what this can churn out in the next few years 🚀
Excited to see what this can churn out in the next few years 🚀
Might just be the best package I've ever received from a collaborator. God bless the Belgians! 🇧🇪🍻

February 12, 2025 at 1:52 PM
Might just be the best package I've ever received from a collaborator. God bless the Belgians! 🇧🇪🍻
There's nothing quite like the excitement of seeing 2D classes of a protein for the first time—but witnessing the first CPE of a virus your lab is just starting to work on is equally satisfying! 🦠

January 17, 2025 at 7:28 AM
There's nothing quite like the excitement of seeing 2D classes of a protein for the first time—but witnessing the first CPE of a virus your lab is just starting to work on is equally satisfying! 🦠
Had a great day at the 10th anniversary Science for Life meeting. My personal highlights were the incredible #cryoET talk from Julia Mahamid, and @bobcoelingh.bsky.social winning a poster prize! 🏆


December 2, 2024 at 5:58 PM
Had a great day at the 10th anniversary Science for Life meeting. My personal highlights were the incredible #cryoET talk from Julia Mahamid, and @bobcoelingh.bsky.social winning a poster prize! 🏆
So it begins...

December 1, 2024 at 1:12 PM
So it begins...
Greatly enjoying our ongoing #cryoET adventures. My new favourite way to procrastinate from administrative tasks is to play I spy with @bobcoelingh.bsky.social's recent data set! 👀
Here's an easy one! 👇
Here's an easy one! 👇

November 30, 2024 at 11:09 AM
Greatly enjoying our ongoing #cryoET adventures. My new favourite way to procrastinate from administrative tasks is to play I spy with @bobcoelingh.bsky.social's recent data set! 👀
Here's an easy one! 👇
Here's an easy one! 👇

