@dandergassen.bsky.social
Independent Junior Group Leader at TUM - Postdoc in the Rinn & Meissner Lab (Harvard) - PhD in Denise Barlow's lab (CeMM)
This work, led by Lison Lemoine and co-authors @sarahhoelzl.bsky.social, @hasenbeint.bsky.social and Elisabeth Graf @tum.de, has been published today in Scientific Reports: www.nature.com/articles/s41...

November 11, 2025 at 7:00 PM
This work, led by Lison Lemoine and co-authors @sarahhoelzl.bsky.social, @hasenbeint.bsky.social and Elisabeth Graf @tum.de, has been published today in Scientific Reports: www.nature.com/articles/s41...
This workflow provides a powerful and accessible framework for studying allele-specific transcript diversity, highlighting the mechanistic insights made possible by long-read transcriptomic data.
November 11, 2025 at 7:00 PM
This workflow provides a powerful and accessible framework for studying allele-specific transcript diversity, highlighting the mechanistic insights made possible by long-read transcriptomic data.
We explored imprinted loci, known for allele-specific coding and non-coding isoforms, and successfully benchmarked historical findings. Our approach also uncovered isoforms expressed from both the active and inactive X chromosomes of escape genes in females.
November 11, 2025 at 7:00 PM
We explored imprinted loci, known for allele-specific coding and non-coding isoforms, and successfully benchmarked historical findings. Our approach also uncovered isoforms expressed from both the active and inactive X chromosomes of escape genes in females.
Thank you 🔥
October 25, 2025 at 9:57 AM
Thank you 🔥
All mouse (6 major organs) and human (54 GTEx tissues) lncRNA-to-target predictions are easily accessible for visual inspection, allowing researchers to select candidates based on target and mechanism predictions
GitHub IGV resource link: github.com/AndergassenL...
GitHub IGV resource link: github.com/AndergassenL...

Allelome.LINK/02_resource at main · AndergassenLab/Allelome.LINK
Contribute to AndergassenLab/Allelome.LINK development by creating an account on GitHub.
github.com
October 24, 2025 at 2:44 AM
All mouse (6 major organs) and human (54 GTEx tissues) lncRNA-to-target predictions are easily accessible for visual inspection, allowing researchers to select candidates based on target and mechanism predictions
GitHub IGV resource link: github.com/AndergassenL...
GitHub IGV resource link: github.com/AndergassenL...
Importantly, as more individual sequencing data and risk variants become available, we anticipate that this strategy will continue to elucidate targets and mechanisms, ultimately decoding the entire cis-acting landscape of the non-coding genome.
October 24, 2025 at 2:44 AM
Importantly, as more individual sequencing data and risk variants become available, we anticipate that this strategy will continue to elucidate targets and mechanisms, ultimately decoding the entire cis-acting landscape of the non-coding genome.
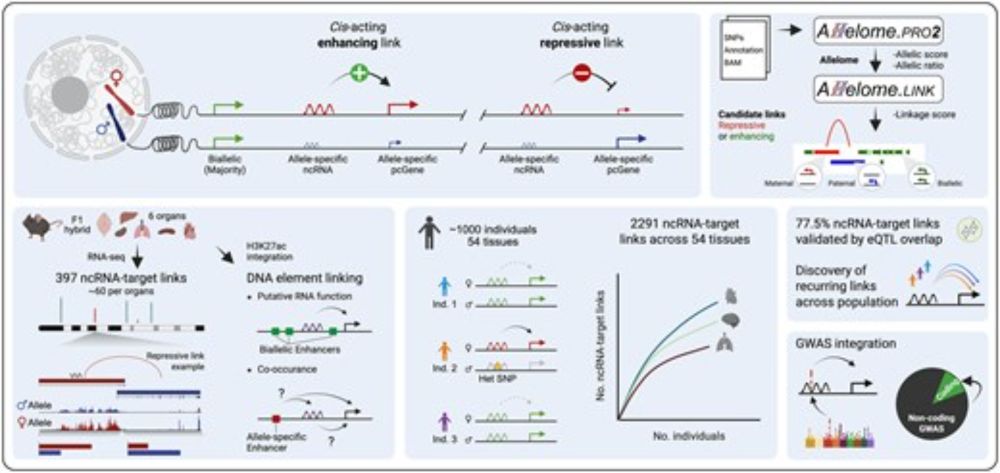
We applied the strategy to mouse organs and to the largest allele-specific human dataset @gtexportal.bsky.social, including 54 tissues from nearly 1000 individuals. Given the high genetic diversity in humans, each individual allows for the discovery of new allelic correlation events.
October 24, 2025 at 2:44 AM
We applied the strategy to mouse organs and to the largest allele-specific human dataset @gtexportal.bsky.social, including 54 tissues from nearly 1000 individuals. Given the high genetic diversity in humans, each individual allows for the discovery of new allelic correlation events.
By further integrating H3K27ac data, we showed that the same principle can be used to link enhancers and other cis-acting DNA regulatory elements to their corresponding targets!
October 24, 2025 at 2:44 AM
By further integrating H3K27ac data, we showed that the same principle can be used to link enhancers and other cis-acting DNA regulatory elements to their corresponding targets!
The strategy is simple: A repressive lncRNA-to-target prediction occurs when the lncRNA and nearby protein-coding genes show opposite allele-specific expression biases, while an enhancing interaction occurs when both show allelic expression bias toward the same allele.
October 24, 2025 at 2:44 AM
The strategy is simple: A repressive lncRNA-to-target prediction occurs when the lncRNA and nearby protein-coding genes show opposite allele-specific expression biases, while an enhancing interaction occurs when both show allelic expression bias toward the same allele.
This work, led by @hasenbeint.bsky.social and co-authors @sarahhoelzl.bsky.social, Stefan Engelhardt at the @tum.de #TRR267, has been published in @narjournal.bsky.social
link: doi.org/10.1093/nar/...
link: doi.org/10.1093/nar/...
Allele-specific genomics decodes gene targets and mechanisms of the non-coding genome
Abstract. A large proportion of disease variants is found in non-coding RNAs (ncRNAs), gene loci that have been identified as key regulatory elements. Howe
doi.org
October 24, 2025 at 2:44 AM
This work, led by @hasenbeint.bsky.social and co-authors @sarahhoelzl.bsky.social, Stefan Engelhardt at the @tum.de #TRR267, has been published in @narjournal.bsky.social
link: doi.org/10.1093/nar/...
link: doi.org/10.1093/nar/...
Yes! The Allelome.LINK framework integrates allele-specific transcriptomics and/or epigenomics to connect cis-acting lncRNAs and enhancers with their nearby protein-coding targets, thereby linking overlapping non-coding disease variants to the genes they affect.
October 24, 2025 at 2:44 AM
Yes! The Allelome.LINK framework integrates allele-specific transcriptomics and/or epigenomics to connect cis-acting lncRNAs and enhancers with their nearby protein-coding targets, thereby linking overlapping non-coding disease variants to the genes they affect.
Finally, we would like to thank all the reviewers for their valuable comments, the @nataging.nature.com editors, as well as Anton Wutz for highlighting our research in the News & Views section of Nature Aging: rdcu.be/eknAp

X inactivation shows frail ends when mice age
Nature Aging - Most genes on the inactive X chromosome are repressed throughout life, but there are exceptions. Hoelzl and colleagues map genes that escape repression in multiple organs over the...
rdcu.be
May 2, 2025 at 7:57 PM
Finally, we would like to thank all the reviewers for their valuable comments, the @nataging.nature.com editors, as well as Anton Wutz for highlighting our research in the News & Views section of Nature Aging: rdcu.be/eknAp
Overall, the escape landscape shows a high degree of organ and cell type specificity, with strong evidence of cluster organization. Explore the full Escape Atlas in the IGV browser for each organ and age time point: github.com/AndergassenL...

AgingX/02_resource at main · AndergassenLab/AgingX
Contribute to AndergassenLab/AgingX development by creating an account on GitHub.
github.com
May 2, 2025 at 7:57 PM
Overall, the escape landscape shows a high degree of organ and cell type specificity, with strong evidence of cluster organization. Explore the full Escape Atlas in the IGV browser for each organ and age time point: github.com/AndergassenL...
Since we find that several age-specific escapees are associated with human diseases, we propose that their elevated expression in females may contribute to sex-biased disease progression with age, offering a new mechanism for age-related sex differences beyond hormonal influence.
May 2, 2025 at 7:57 PM
Since we find that several age-specific escapees are associated with human diseases, we propose that their elevated expression in females may contribute to sex-biased disease progression with age, offering a new mechanism for age-related sex differences beyond hormonal influence.
In addition, allele-specific single-cell analysis revealed that age-specific escape manifests within distinct cell types, further providing evidence that age-related epigenetic changes promote gene escape.

May 2, 2025 at 7:57 PM
In addition, allele-specific single-cell analysis revealed that age-specific escape manifests within distinct cell types, further providing evidence that age-related epigenetic changes promote gene escape.

