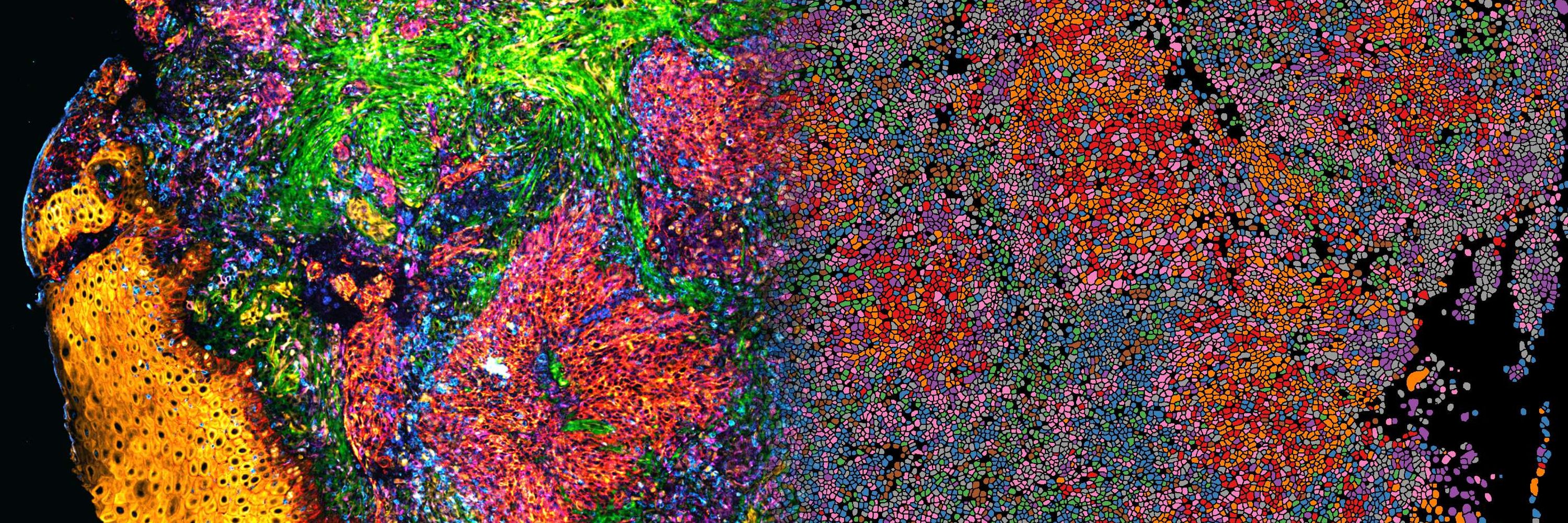
– A simplified fast-track pipeline for easy adoption
– An AI-powered option for advanced image & spatial analysis
👉 BioRxiv: bit.ly/44SBuHu
👉 Github: bit.ly/4eR3IqK

– A simplified fast-track pipeline for easy adoption
– An AI-powered option for advanced image & spatial analysis
👉 BioRxiv: bit.ly/44SBuHu
👉 Github: bit.ly/4eR3IqK
Kudos to @melissaklingeberg.bsky.social 🥳 and big thanks to all our collaborators! bit.ly/3SCARw4

Kudos to @melissaklingeberg.bsky.social 🥳 and big thanks to all our collaborators! bit.ly/3SCARw4
We demonstrate how ultrasensitive discovery proteomics can shed light onto early carcinogenesis, uncover cell-of-origin signatures, and prioritize novel drug targets.
bit.ly/41M7fke

We demonstrate how ultrasensitive discovery proteomics can shed light onto early carcinogenesis, uncover cell-of-origin signatures, and prioritize novel drug targets.
bit.ly/41M7fke
and the speakers @bonalditiziana.bsky.social, @coscialab.bsky.social, @kusterlab.bsky.social, @anamdv.bsky.social, @jsandow.bsky.social

and the speakers @bonalditiziana.bsky.social, @coscialab.bsky.social, @kusterlab.bsky.social, @anamdv.bsky.social, @jsandow.bsky.social
Georg Kustacher - bioinformatics
@coscialab.bsky.social single cell/spatial session
Manuel Mayr - clinical proteomics
Richard Seltema - structural
Freddie Theodolou - non-human proteomics
Jyoti Choudary - functional proteomics
Georg Kustacher - bioinformatics
@coscialab.bsky.social single cell/spatial session
Manuel Mayr - clinical proteomics
Richard Seltema - structural
Freddie Theodolou - non-human proteomics
Jyoti Choudary - functional proteomics
Keynote Speakers:
@shalinhnaik.bsky.social
Pulin Li, and me!
Invited Speakers:
@emmanuel-saliba.bsky.social
@mratz.bsky.social
Barbara Grünwald
Maria Colomé-Tatché
Mohammad Lotfollahi
@coscialab.bsky.social
Teresa Krieger
www.isco-conference.eu

Keynote Speakers:
@shalinhnaik.bsky.social
Pulin Li, and me!
Invited Speakers:
@emmanuel-saliba.bsky.social
@mratz.bsky.social
Barbara Grünwald
Maria Colomé-Tatché
Mohammad Lotfollahi
@coscialab.bsky.social
Teresa Krieger
www.isco-conference.eu




