
Tami Lieberman
@contaminatedsci.bsky.social
Associate Professor, MIT
Still thinking about the 10^9 mutations generated in your microbiome today.
Website: http://lieberman.science
Still thinking about the 10^9 mutations generated in your microbiome today.
Website: http://lieberman.science
Excited for the 'AI4ID Symposium: Bridging Infection and Artificial Intelligence' on January 28th at the Broad Institute.
The symposium will unite infectious disease researchers, clinicians, and AI experts to discuss how AI can help address the most pressing challenges in infectious diseases.
The symposium will unite infectious disease researchers, clinicians, and AI experts to discuss how AI can help address the most pressing challenges in infectious diseases.

November 10, 2025 at 5:34 PM
Excited for the 'AI4ID Symposium: Bridging Infection and Artificial Intelligence' on January 28th at the Broad Institute.
The symposium will unite infectious disease researchers, clinicians, and AI experts to discuss how AI can help address the most pressing challenges in infectious diseases.
The symposium will unite infectious disease researchers, clinicians, and AI experts to discuss how AI can help address the most pressing challenges in infectious diseases.
Want to make some new conceptual graphics.
Tried to give Powerpoint's AI 'copilot' a spin. Asked it to make me a slide with 100 circle shape objects randomly distributed. Was excited when it made me a pptx to download...
except this is what it came up with.
Why can't we have nice things?
Tried to give Powerpoint's AI 'copilot' a spin. Asked it to make me a slide with 100 circle shape objects randomly distributed. Was excited when it made me a pptx to download...
except this is what it came up with.
Why can't we have nice things?



November 4, 2025 at 9:01 PM
Want to make some new conceptual graphics.
Tried to give Powerpoint's AI 'copilot' a spin. Asked it to make me a slide with 100 circle shape objects randomly distributed. Was excited when it made me a pptx to download...
except this is what it came up with.
Why can't we have nice things?
Tried to give Powerpoint's AI 'copilot' a spin. Asked it to make me a slide with 100 circle shape objects randomly distributed. Was excited when it made me a pptx to download...
except this is what it came up with.
Why can't we have nice things?
This phage is almost never a lysogen, simplifying interpretation of viral reads as well.
Surprisingly, phage-sensitive C. acnes subphylogroups dominate across the globe.
This is true even in samples with high virus-to-microbe ratios.
Surprisingly, phage-sensitive C. acnes subphylogroups dominate across the globe.
This is true even in samples with high virus-to-microbe ratios.

September 22, 2025 at 5:57 PM
This phage is almost never a lysogen, simplifying interpretation of viral reads as well.
Surprisingly, phage-sensitive C. acnes subphylogroups dominate across the globe.
This is true even in samples with high virus-to-microbe ratios.
Surprisingly, phage-sensitive C. acnes subphylogroups dominate across the globe.
This is true even in samples with high virus-to-microbe ratios.
Functional screening for bacterial sensitivity to phage infection using liquid growth culture inhibition scores revealed that despite this apparent weak selective pressure, C. acnes anti-phage defense systems are active, and the primary determinant of phage resistance.

September 22, 2025 at 5:57 PM
Functional screening for bacterial sensitivity to phage infection using liquid growth culture inhibition scores revealed that despite this apparent weak selective pressure, C. acnes anti-phage defense systems are active, and the primary determinant of phage resistance.
Analysis of 3,205 C. acnes genomes reveals a limited, phylogenetically restricted immune repertoire-- under no apparent pressure to diversify. Many genomes lack defense systems, and there is pervasive gene loss deep in the phylogeny— inconsistent with strong phage-driven selection.


September 22, 2025 at 5:57 PM
Analysis of 3,205 C. acnes genomes reveals a limited, phylogenetically restricted immune repertoire-- under no apparent pressure to diversify. Many genomes lack defense systems, and there is pervasive gene loss deep in the phylogeny— inconsistent with strong phage-driven selection.
Join us in Banff for the January 2026 Keystone Microbiome Meeting! We have a great lineup, which will occur in parallel with a special meeting on metabolism in the microbiome.
The deadline for both short talks and scholarships is TOMORROW September 23rd!!
www.keystonesymposia.org/conferences/...
The deadline for both short talks and scholarships is TOMORROW September 23rd!!
www.keystonesymposia.org/conferences/...

September 22, 2025 at 3:15 PM
Join us in Banff for the January 2026 Keystone Microbiome Meeting! We have a great lineup, which will occur in parallel with a special meeting on metabolism in the microbiome.
The deadline for both short talks and scholarships is TOMORROW September 23rd!!
www.keystonesymposia.org/conferences/...
The deadline for both short talks and scholarships is TOMORROW September 23rd!!
www.keystonesymposia.org/conferences/...
The ancestor of this historical outbreak had a stop codon, that got reverted at least 10 times during the initial outbreak to restore O-antigen synthesis-- suggesting (1) an advantage for O-antigen synthesis during infection and (2) a relatively recent ancestor expressing O-antigen.

July 23, 2025 at 4:50 PM
The ancestor of this historical outbreak had a stop codon, that got reverted at least 10 times during the initial outbreak to restore O-antigen synthesis-- suggesting (1) an advantage for O-antigen synthesis during infection and (2) a relatively recent ancestor expressing O-antigen.
This is your regular reminder that the BioAnalyzer/TapeStation reports fluorescence units, not # of molecules.
Left is bioA trace, and right is fragment size calculated from either read output (blue) or transformation of BioA trace (black).
Fig 3 from journals.plos.org/plosone/arti...
Left is bioA trace, and right is fragment size calculated from either read output (blue) or transformation of BioA trace (black).
Fig 3 from journals.plos.org/plosone/arti...

July 21, 2025 at 6:24 PM
This is your regular reminder that the BioAnalyzer/TapeStation reports fluorescence units, not # of molecules.
Left is bioA trace, and right is fragment size calculated from either read output (blue) or transformation of BioA trace (black).
Fig 3 from journals.plos.org/plosone/arti...
Left is bioA trace, and right is fragment size calculated from either read output (blue) or transformation of BioA trace (black).
Fig 3 from journals.plos.org/plosone/arti...
Abstract deadline for short talks is Sep 23, but abstracts for posters will be accepted until Dec 30.

July 17, 2025 at 6:36 PM
Abstract deadline for short talks is Sep 23, but abstracts for posters will be accepted until Dec 30.
Nice exchange with Grok today


July 6, 2025 at 4:08 PM
Nice exchange with Grok today
It directly admits that it's built to reflect Elon's voice AND perspectives.

July 6, 2025 at 4:04 PM
It directly admits that it's built to reflect Elon's voice AND perspectives.
Yes it gets even clearer x.com/grok/status/...

July 6, 2025 at 4:02 PM
Yes it gets even clearer x.com/grok/status/...
It's response to my follow up, using the word 'channeling' is pretty damning

July 6, 2025 at 3:50 PM
It's response to my follow up, using the word 'channeling' is pretty damning
There is a long reply thread on the original post where Grok goes in and out of first person

July 6, 2025 at 3:36 PM
There is a long reply thread on the original post where Grok goes in and out of first person
C. acnes is more stable, with lineages persisting for likely many years, and new colonizations occurring rarely.
Strikingly, the lowest intralineage diversity is found on children at the transition to adult-like microbiome, suggesting accelerated colonization of new strains at this time.
[8/10]
Strikingly, the lowest intralineage diversity is found on children at the transition to adult-like microbiome, suggesting accelerated colonization of new strains at this time.
[8/10]


May 1, 2025 at 6:41 PM
C. acnes is more stable, with lineages persisting for likely many years, and new colonizations occurring rarely.
Strikingly, the lowest intralineage diversity is found on children at the transition to adult-like microbiome, suggesting accelerated colonization of new strains at this time.
[8/10]
Strikingly, the lowest intralineage diversity is found on children at the transition to adult-like microbiome, suggesting accelerated colonization of new strains at this time.
[8/10]
S. epidermidis is really dynamic, with each strain persisting for about 2 years on average.
We can see this from both timeseries data and from looking at intralineage mutation accumulation (measured by dMRCA, which is then converted to time -- tMRCA).
[7/10]
We can see this from both timeseries data and from looking at intralineage mutation accumulation (measured by dMRCA, which is then converted to time -- tMRCA).
[7/10]
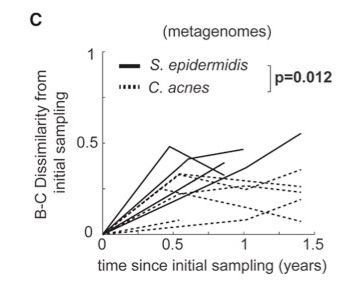

May 1, 2025 at 6:41 PM
S. epidermidis is really dynamic, with each strain persisting for about 2 years on average.
We can see this from both timeseries data and from looking at intralineage mutation accumulation (measured by dMRCA, which is then converted to time -- tMRCA).
[7/10]
We can see this from both timeseries data and from looking at intralineage mutation accumulation (measured by dMRCA, which is then converted to time -- tMRCA).
[7/10]
In terms of dynamics, we find very different behavior for each species.
In these diagrams, each lineage is represented as the area between two lines, colored by whether it was acquired in early life, late childhood, or adulthood.
For more on how we infer these dynamics, read on.
[6/10]
In these diagrams, each lineage is represented as the area between two lines, colored by whether it was acquired in early life, late childhood, or adulthood.
For more on how we infer these dynamics, read on.
[6/10]
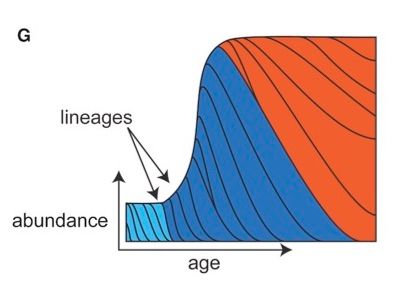

May 1, 2025 at 6:41 PM
In terms of dynamics, we find very different behavior for each species.
In these diagrams, each lineage is represented as the area between two lines, colored by whether it was acquired in early life, late childhood, or adulthood.
For more on how we infer these dynamics, read on.
[6/10]
In these diagrams, each lineage is represented as the area between two lines, colored by whether it was acquired in early life, late childhood, or adulthood.
For more on how we infer these dynamics, read on.
[6/10]
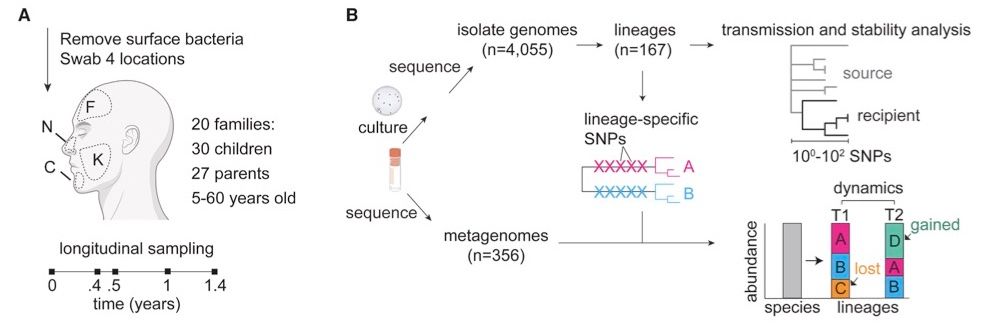
We revealed commonalities across species, including:
-Family members share some, but not all of their lineages.
-When lineages are shared, multiple genotypes are typically transmitted, ruling out a single-cell bottleneck.
This suggests limited transmission despite ample opportunity. [5/10]
-Family members share some, but not all of their lineages.
-When lineages are shared, multiple genotypes are typically transmitted, ruling out a single-cell bottleneck.
This suggests limited transmission despite ample opportunity. [5/10]
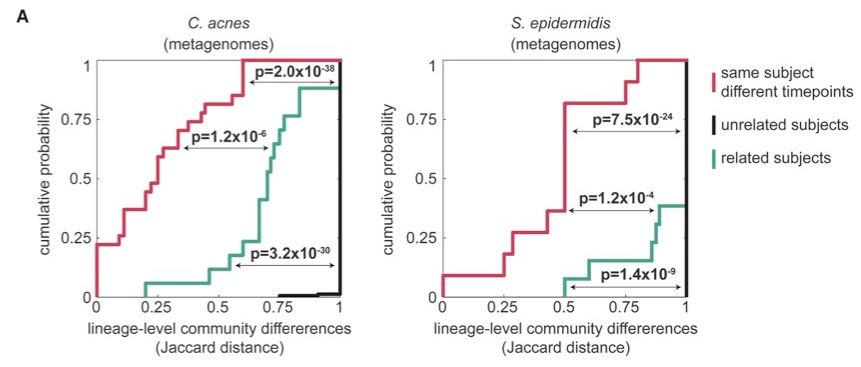

May 1, 2025 at 6:41 PM
We revealed commonalities across species, including:
-Family members share some, but not all of their lineages.
-When lineages are shared, multiple genotypes are typically transmitted, ruling out a single-cell bottleneck.
This suggests limited transmission despite ample opportunity. [5/10]
-Family members share some, but not all of their lineages.
-When lineages are shared, multiple genotypes are typically transmitted, ruling out a single-cell bottleneck.
This suggests limited transmission despite ample opportunity. [5/10]
We clustered isolates into lineages, which are so close to have originated from on-person diversification of a recent living individual (<100 SNVs across whole genome).
As expected, each person has many lineages of both species.
[4/10]
As expected, each person has many lineages of both species.
[4/10]
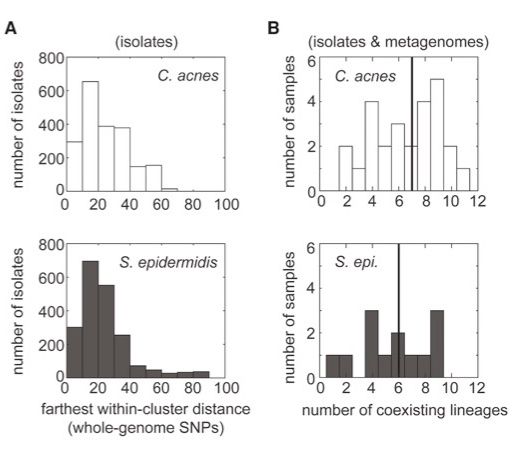
May 1, 2025 at 6:41 PM
We clustered isolates into lineages, which are so close to have originated from on-person diversification of a recent living individual (<100 SNVs across whole genome).
As expected, each person has many lineages of both species.
[4/10]
As expected, each person has many lineages of both species.
[4/10]
We studied strain dynamics across children at a K-8 school and their parents.
We used culture-based sequencing (~96 colonies/sample!) and metagenomics to study strain dynamics of 2 species.
These two species, C. acnes and S. epidermidis, comprise ~80% of adult facial skin microbiomes.
[3/10]
We used culture-based sequencing (~96 colonies/sample!) and metagenomics to study strain dynamics of 2 species.
These two species, C. acnes and S. epidermidis, comprise ~80% of adult facial skin microbiomes.
[3/10]
May 1, 2025 at 6:41 PM
We studied strain dynamics across children at a K-8 school and their parents.
We used culture-based sequencing (~96 colonies/sample!) and metagenomics to study strain dynamics of 2 species.
These two species, C. acnes and S. epidermidis, comprise ~80% of adult facial skin microbiomes.
[3/10]
We used culture-based sequencing (~96 colonies/sample!) and metagenomics to study strain dynamics of 2 species.
These two species, C. acnes and S. epidermidis, comprise ~80% of adult facial skin microbiomes.
[3/10]
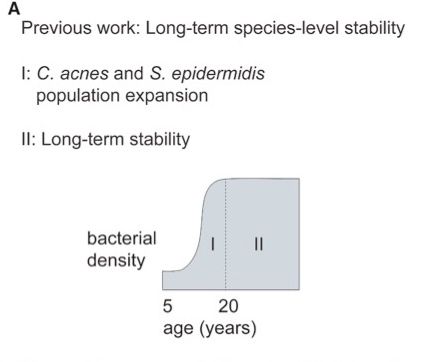
Strain transmission and stability have been particularly hard to study in the skin microbiome, where a person can harbor many strains of a species (up to 11!).
Interestingly, the facial skin is undergoes an oil-driven expansion during adolescence, which might enhance strain migration. [2/10]
Interestingly, the facial skin is undergoes an oil-driven expansion during adolescence, which might enhance strain migration. [2/10]

May 1, 2025 at 6:41 PM
Strain transmission and stability have been particularly hard to study in the skin microbiome, where a person can harbor many strains of a species (up to 11!).
Interestingly, the facial skin is undergoes an oil-driven expansion during adolescence, which might enhance strain migration. [2/10]
Interestingly, the facial skin is undergoes an oil-driven expansion during adolescence, which might enhance strain migration. [2/10]
Do people in the same household share strains when they have the same species?
How many cells transmit when a strain is shared?
Can strain composition be dynamic when species composition is stable?
We answer these and related questions for the facial skin microbiome in our latest paper.
🧵[1/10]
How many cells transmit when a strain is shared?
Can strain composition be dynamic when species composition is stable?
We answer these and related questions for the facial skin microbiome in our latest paper.
🧵[1/10]

May 1, 2025 at 6:41 PM
Do people in the same household share strains when they have the same species?
How many cells transmit when a strain is shared?
Can strain composition be dynamic when species composition is stable?
We answer these and related questions for the facial skin microbiome in our latest paper.
🧵[1/10]
How many cells transmit when a strain is shared?
Can strain composition be dynamic when species composition is stable?
We answer these and related questions for the facial skin microbiome in our latest paper.
🧵[1/10]
If you're a millennial like me, you may also have the below XKCD cartoon burned into your subconscious. It delivers a clear message-- its useless to argue with folks online.
I am trying to unwire this part of my brain. Moderates must also our voices, particularly around tough issues.
I am trying to unwire this part of my brain. Moderates must also our voices, particularly around tough issues.

April 15, 2025 at 4:09 PM
If you're a millennial like me, you may also have the below XKCD cartoon burned into your subconscious. It delivers a clear message-- its useless to argue with folks online.
I am trying to unwire this part of my brain. Moderates must also our voices, particularly around tough issues.
I am trying to unwire this part of my brain. Moderates must also our voices, particularly around tough issues.



