
STORIES learns cell fate landscapes from spatial tramscripromics data profiled at several time points, thus allowing prediction of future cell states.
Led by Geert-Jan Huizing and Jules Samaran
www.nature.com/articles/s41...
@pasteur.fr

STORIES learns cell fate landscapes from spatial tramscripromics data profiled at several time points, thus allowing prediction of future cell states.
Led by Geert-Jan Huizing and Jules Samaran
www.nature.com/articles/s41...
@pasteur.fr
Based on #Cicero 's algorithm (Pliner et al.), it runs ~150x faster, processing an atlas of 700k cells in less than 40 min! ⛷️
Short paper: doi.org/10.1101/2025...
Code: github.com/cantinilab/CIRCE
1/5 ⬇️

Based on #Cicero 's algorithm (Pliner et al.), it runs ~150x faster, processing an atlas of 700k cells in less than 40 min! ⛷️
Short paper: doi.org/10.1101/2025...
Code: github.com/cantinilab/CIRCE
1/5 ⬇️
Our #ICLR2025 blog post on Flow Matching was published yesterday : iclr-blogposts.github.io/2025/blog/co...
My PhD student @annegnx.bsky.social will present it tomorrow in ICLR, 👉poster session 4, 3 pm, #549 in Hall 3/2B 👈
Our #ICLR2025 blog post on Flow Matching was published yesterday : iclr-blogposts.github.io/2025/blog/co...
My PhD student @annegnx.bsky.social will present it tomorrow in ICLR, 👉poster session 4, 3 pm, #549 in Hall 3/2B 👈
www.nature.com/artic...
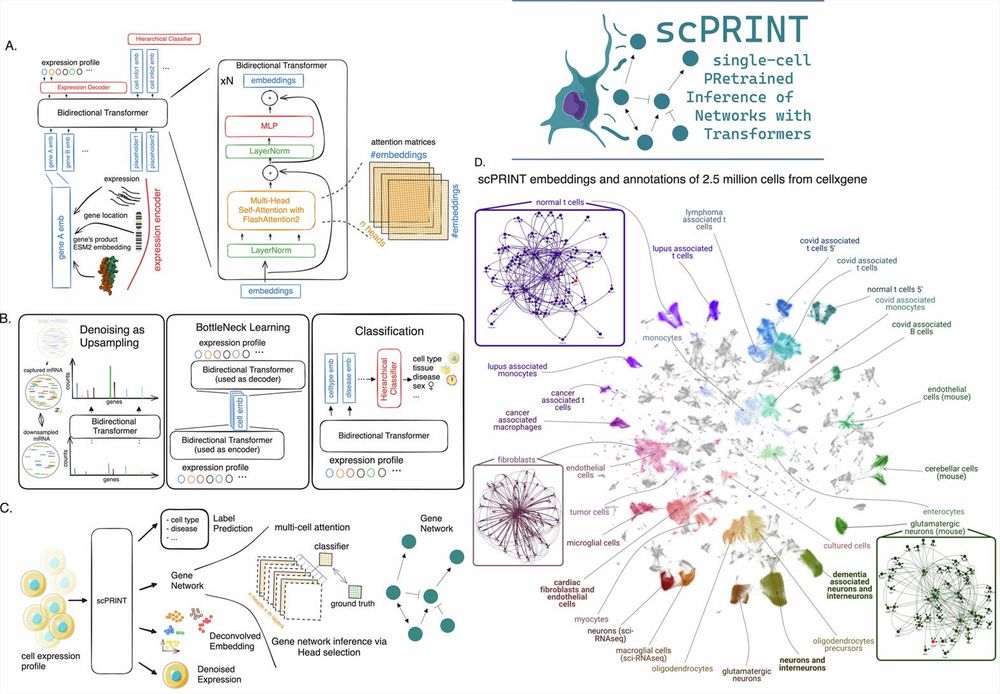
www.nature.com/artic...

www.nature.com/artic...
@m-albert.bsky.social is now teaching AI for image analysis. Felipe Llinares from Bioptimus will then talk about LLMs, with short talks from @jkobject.com from my team and @emordret.bsky.social from @audeber.bsky.social team.
@pasteur.fr @pasteuredu.bsky.social

@m-albert.bsky.social is now teaching AI for image analysis. Felipe Llinares from Bioptimus will then talk about LLMs, with short talks from @jkobject.com from my team and @emordret.bsky.social from @audeber.bsky.social team.
@pasteur.fr @pasteuredu.bsky.social
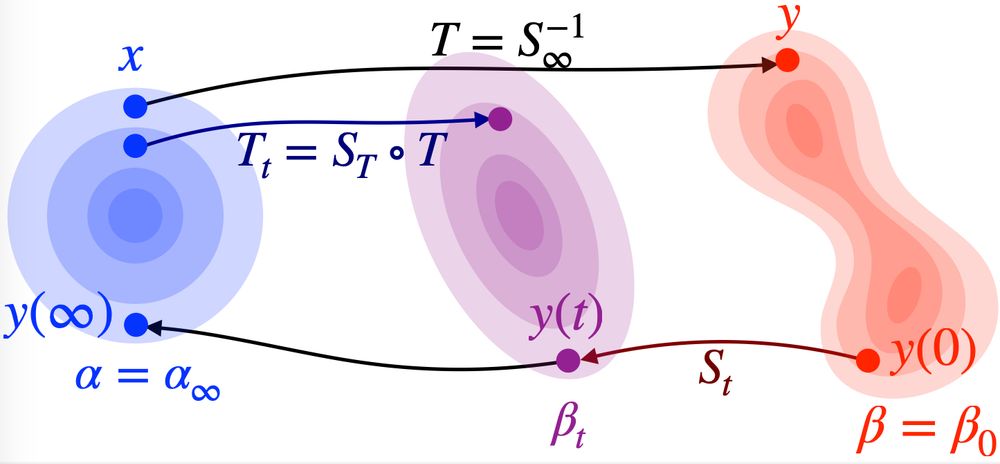
We got this idea after their cool work on improving Plug and Play with FM: arxiv.org/abs/2410.02423
We got this idea after their cool work on improving Plug and Play with FM: arxiv.org/abs/2410.02423
it is possible to embed any cloud of N points from R^d into R^k without distorting their respective distances too much, provided k is not too small (independently of d!)
Better: any random Gaussian embedding works with high proba!
it is possible to embed any cloud of N points from R^d into R^k without distorting their respective distances too much, provided k is not too small (independently of d!)
Better: any random Gaussian embedding works with high proba!
Come join the Cantinilab!
emploi.cnrs.fr/Offres/CDD/U... (better english version coming up soon)
Come join the Cantinilab!
emploi.cnrs.fr/Offres/CDD/U... (better english version coming up soon)

