
ChimeraX
@chimerax.ucsf.edu
UCSF ChimeraX - software for visualizing biomolecules
Here's how to predict binding of tens or hundreds of small molecules to protein assemblies using Boltz 2 in ChimeraX.
Accuracy depends on how similar the ligands and binding pockets are to existing experimental structures.
www.rbvi.ucsf.edu/chimerax/dat...
Accuracy depends on how similar the ligands and binding pockets are to existing experimental structures.
www.rbvi.ucsf.edu/chimerax/dat...
August 20, 2025 at 9:01 PM
Here's how to predict binding of tens or hundreds of small molecules to protein assemblies using Boltz 2 in ChimeraX.
Accuracy depends on how similar the ligands and binding pockets are to existing experimental structures.
www.rbvi.ucsf.edu/chimerax/dat...
Accuracy depends on how similar the ligands and binding pockets are to existing experimental structures.
www.rbvi.ucsf.edu/chimerax/dat...
ChimeraX daily builds can predict binding affinity of small molecules using Boltz 2 on your Mac, Windows or Linux computer. www.rbvi.ucsf.edu/chimerax/dat...
July 24, 2025 at 2:44 AM
ChimeraX daily builds can predict binding affinity of small molecules using Boltz 2 on your Mac, Windows or Linux computer. www.rbvi.ucsf.edu/chimerax/dat...
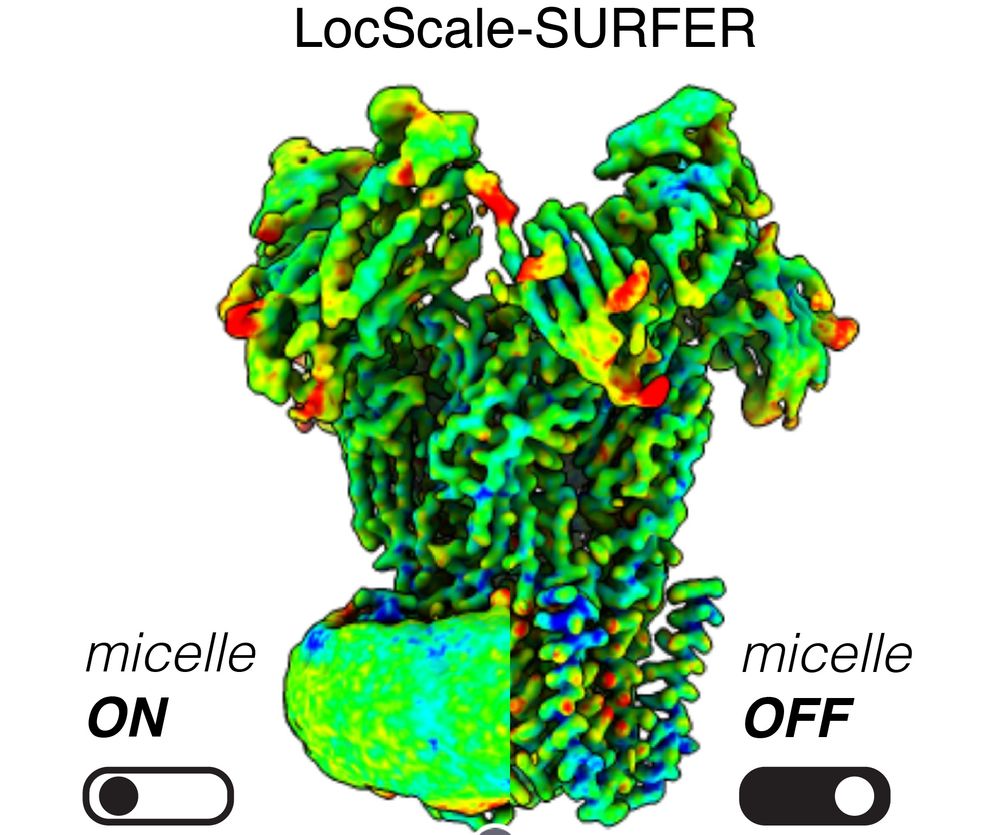
LocScale-SURFER is a ChimeraX extension that shows cryoEM density maps after removing detergent micelles based on output from command-line program Locscale-2.0. Install it using ChimeraX menu Tools / More Tools.... More details at cxtoolshed.rbvi.ucsf.edu/apps/chimera...
June 10, 2025 at 6:12 PM
LocScale-SURFER is a ChimeraX extension that shows cryoEM density maps after removing detergent micelles based on output from command-line program Locscale-2.0. Install it using ChimeraX menu Tools / More Tools.... More details at cxtoolshed.rbvi.ucsf.edu/apps/chimera...
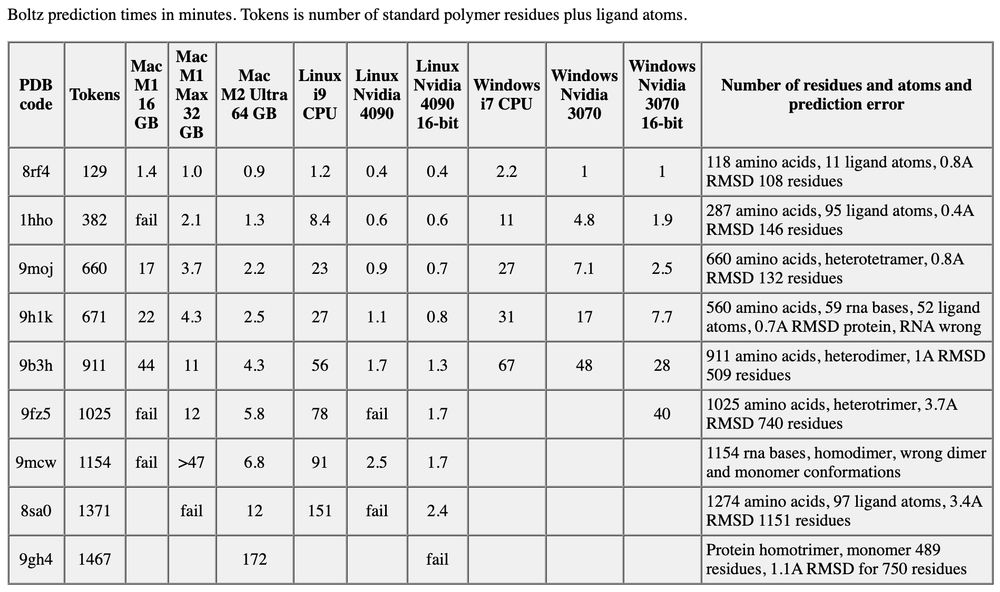
ChimeraX can predict protein, nucleic acid and ligand atomic models on your Mac, Windows or Linux computer using Boltz. This table shows how long it takes in minutes using different computers. www.rbvi.ucsf.edu/chimerax/dat...
May 31, 2025 at 12:25 AM
ChimeraX can predict protein, nucleic acid and ligand atomic models on your Mac, Windows or Linux computer using Boltz. This table shows how long it takes in minutes using different computers. www.rbvi.ucsf.edu/chimerax/dat...
ChimeraX daily builds can predict small complexes of proteins, nucleic acids and small molecules using Boltz on your Mac, Windows or Linux computer without Nvidia graphics. www.rbvi.ucsf.edu/chimerax/dat...
May 1, 2025 at 12:35 AM
ChimeraX daily builds can predict small complexes of proteins, nucleic acids and small molecules using Boltz on your Mac, Windows or Linux computer without Nvidia graphics. www.rbvi.ucsf.edu/chimerax/dat...
We've just been notified that our previous article in Protein Science (2023) is among the 10 most cited papers for that year! You can find 'UCSF ChimeraX: Tools for structure building and analysis' at doi.org/10.1002/pro....

March 19, 2025 at 8:06 PM
We've just been notified that our previous article in Protein Science (2023) is among the 10 most cited papers for that year! You can find 'UCSF ChimeraX: Tools for structure building and analysis' at doi.org/10.1002/pro....
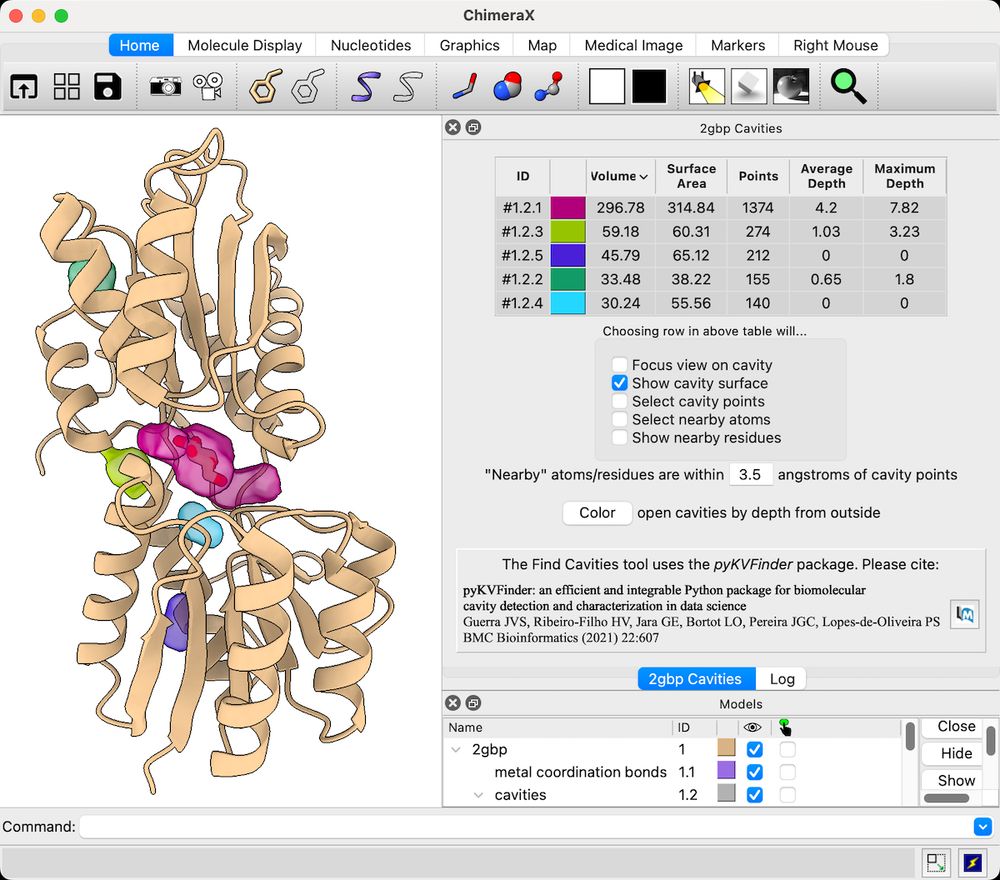
ChimeraX can be used to find pockets and cavities in protein structures using the Find Cavities tool or kvfinder command. This feature is available in daily builds from 10 March 2025 or later.
March 17, 2025 at 8:26 PM
ChimeraX can be used to find pockets and cavities in protein structures using the Find Cavities tool or kvfinder command. This feature is available in daily builds from 10 March 2025 or later.
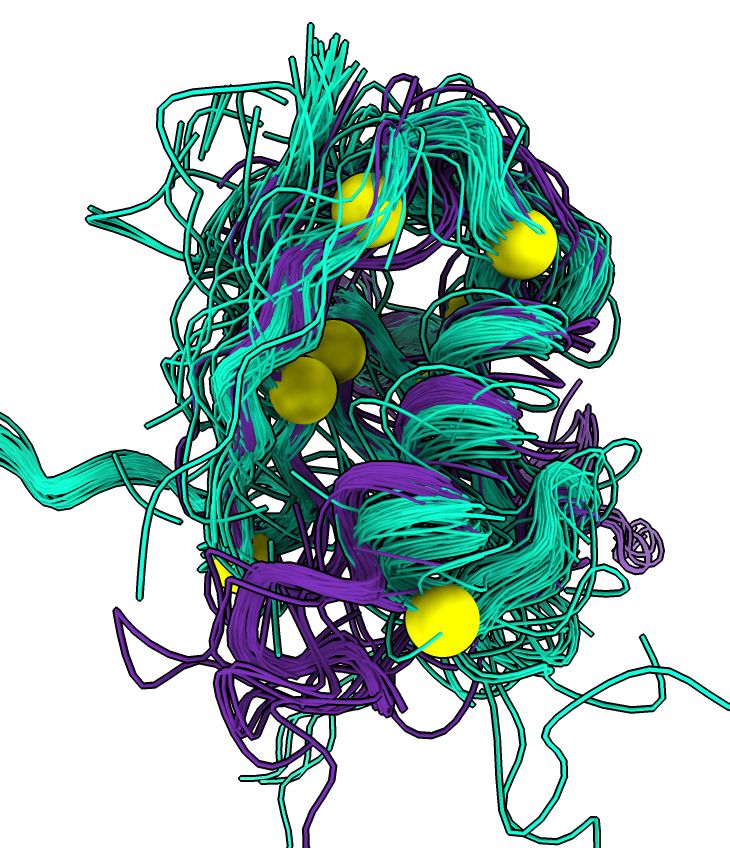
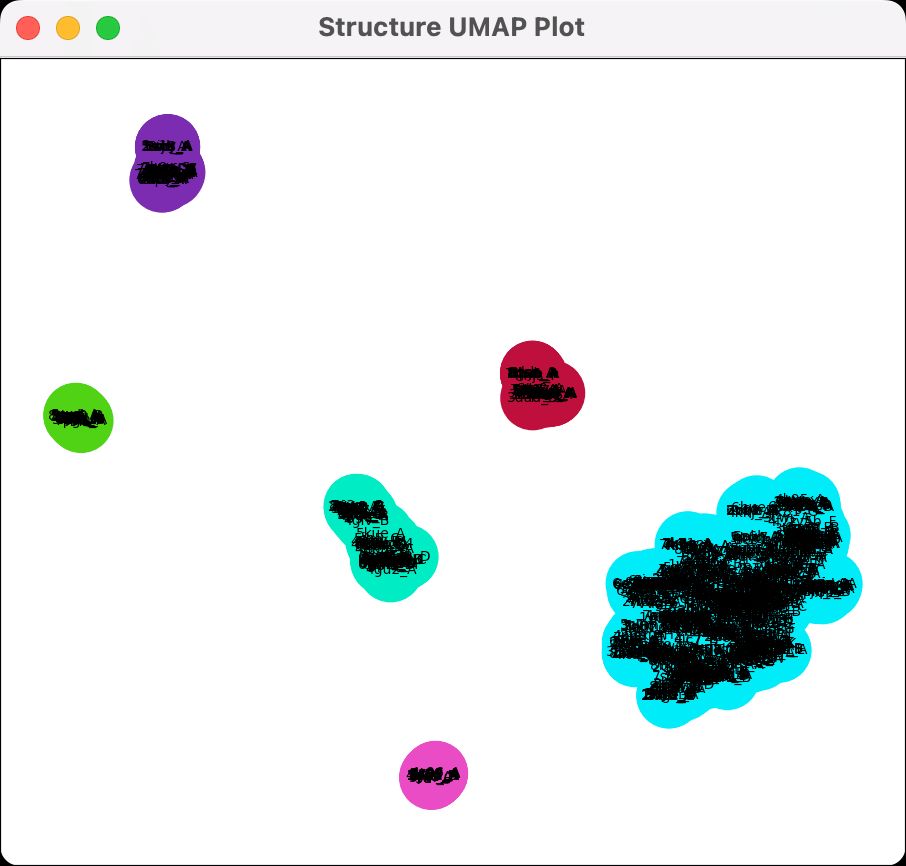
ChimeraX can cluster different backbone conformations of distantly related proteins found with Foldseek, here clustering similar structures to CRAF kinase. www.rbvi.ucsf.edu/chimerax/dat...
March 6, 2025 at 11:25 PM
ChimeraX can cluster different backbone conformations of distantly related proteins found with Foldseek, here clustering similar structures to CRAF kinase. www.rbvi.ucsf.edu/chimerax/dat...
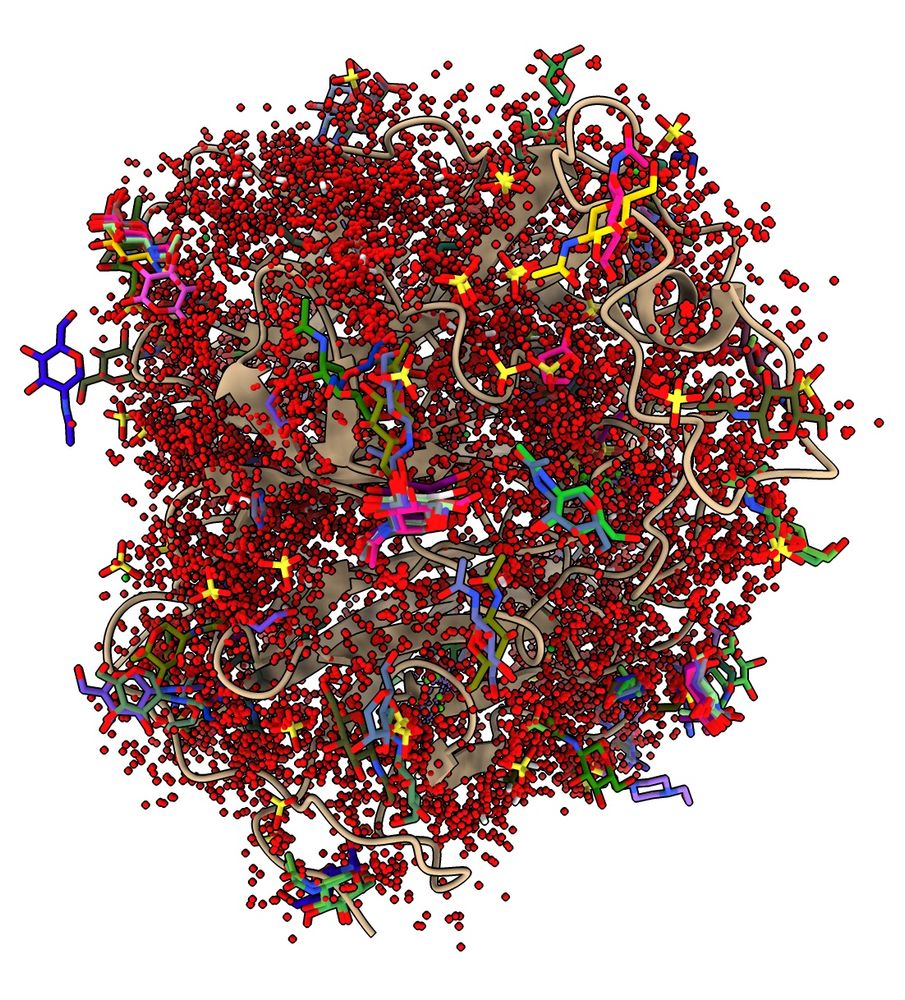
ChimeraX can run Foldseek to find similar structures, such as distantly related homologs, and analyze the results, for example, mapping all ligands onto your query structure. Here are ligands mapped onto Nipah virus G protein. www.rbvi.ucsf.edu/chimerax/dat...
March 6, 2025 at 11:06 PM
ChimeraX can run Foldseek to find similar structures, such as distantly related homologs, and analyze the results, for example, mapping all ligands onto your query structure. Here are ligands mapped onto Nipah virus G protein. www.rbvi.ucsf.edu/chimerax/dat...
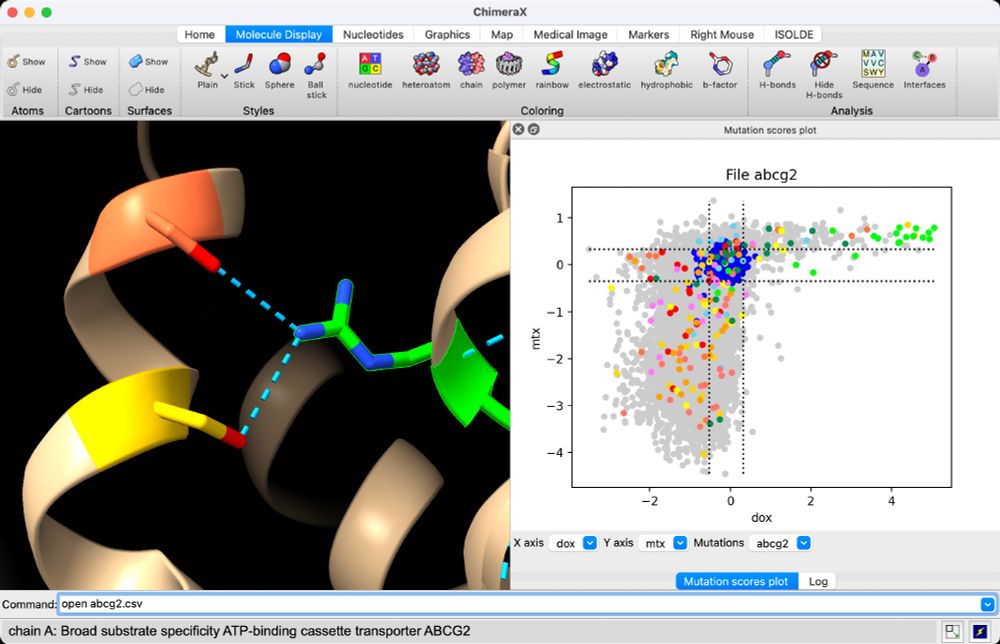
ChimeraX can display deep mutational scan data where all possible single mutations of a protein are assayed using high-throughput experiments. www.rbvi.ucsf.edu/chimerax/dat...
March 6, 2025 at 10:10 PM
ChimeraX can display deep mutational scan data where all possible single mutations of a protein are assayed using high-throughput experiments. www.rbvi.ucsf.edu/chimerax/dat...
Sony Spatial Reality 3D display shows molecular structures and electron microscopy data at HD resolutions with no glasses. www.rbvi.ucsf.edu/chimerax/dat...
March 6, 2025 at 10:01 PM
Sony Spatial Reality 3D display shows molecular structures and electron microscopy data at HD resolutions with no glasses. www.rbvi.ucsf.edu/chimerax/dat...
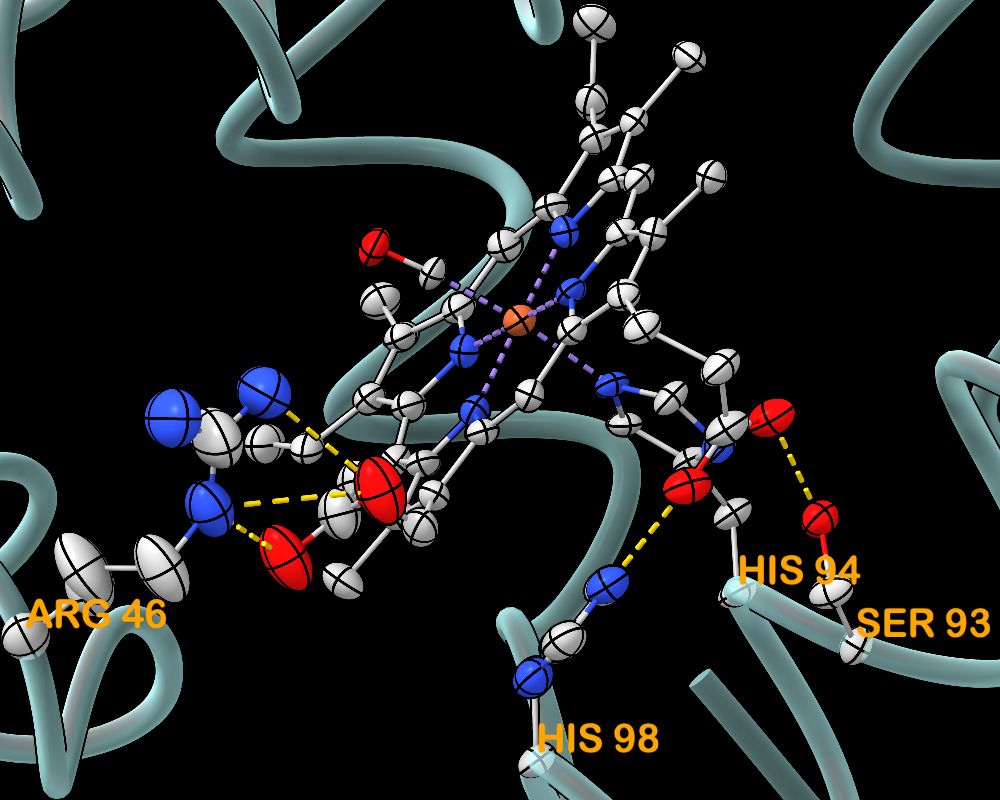
Anisotropic B-factors can now be shown as ellipsoids in ChimeraX daily builds, shown here for the heme in myoglobin. www.rbvi.ucsf.edu/chimerax/fea...
March 6, 2025 at 9:48 PM
Anisotropic B-factors can now be shown as ellipsoids in ChimeraX daily builds, shown here for the heme in myoglobin. www.rbvi.ucsf.edu/chimerax/fea...

