
http://www.dickinsonlab.uchicago.edu/

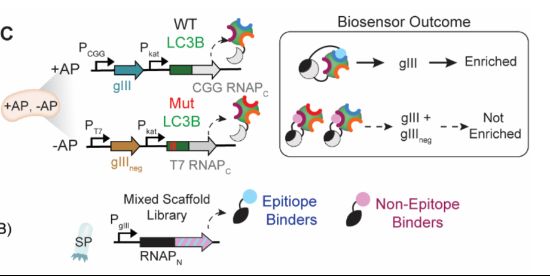
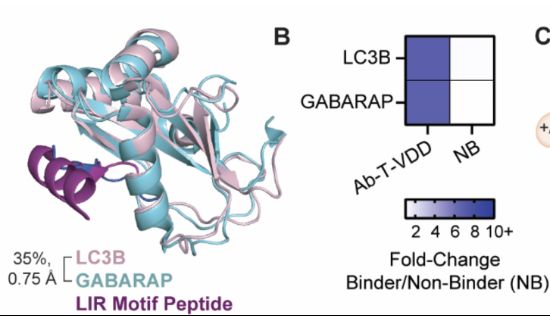



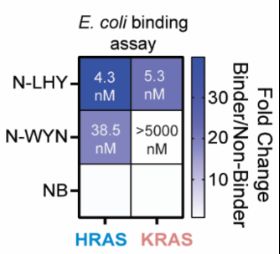













blog.genesmindsmachines.com/p/we-still-c...

blog.genesmindsmachines.com/p/we-still-c...


www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...



