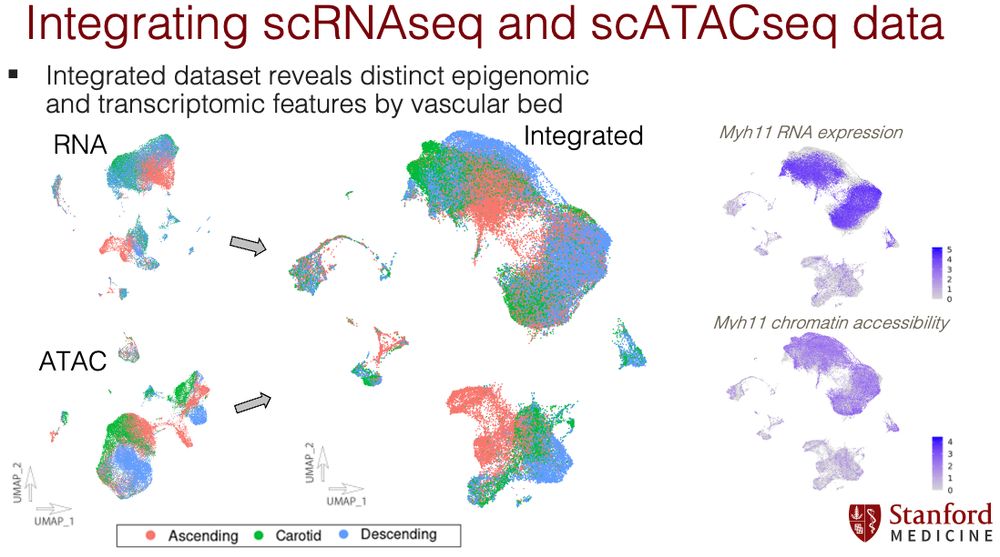
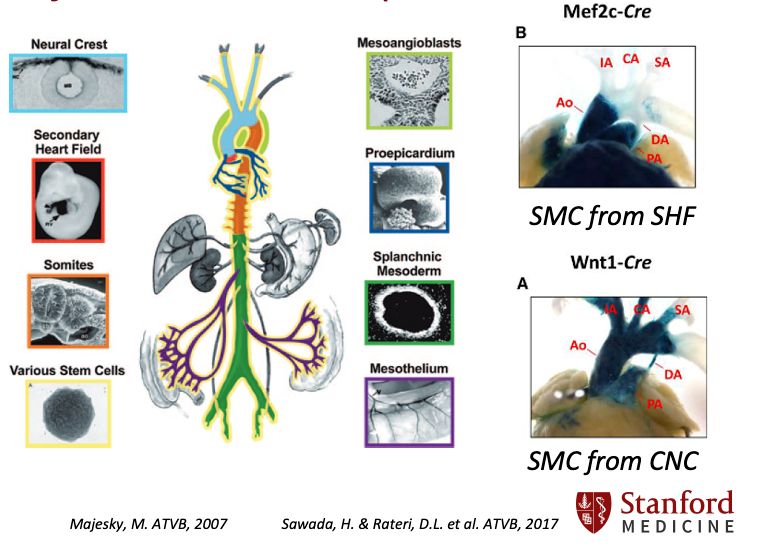
https://profiles.stanford.edu/chad-weldy








We reveal a fundamental observation, that vascular SMC have a unique requirement for ADAR1 editing to prevent MDA5 activation.
SMC deletion of ADAR1 leads to severe phenotype within days and is entirely blocked with deletion of MDA5

We reveal a fundamental observation, that vascular SMC have a unique requirement for ADAR1 editing to prevent MDA5 activation.
SMC deletion of ADAR1 leads to severe phenotype within days and is entirely blocked with deletion of MDA5
was that beyond rare disease, common variants appear to regulate RNA editing (edQTLs), and these edQTLs predict numerous common inflammatory disorders, including CAD! t.co/t1i47lPlGG

was that beyond rare disease, common variants appear to regulate RNA editing (edQTLs), and these edQTLs predict numerous common inflammatory disorders, including CAD! t.co/t1i47lPlGG


So why doesn't this dsRNA induce an antiviral response? ADAR1!

So why doesn't this dsRNA induce an antiviral response? ADAR1!
Even in coral and octopus in response to temperature changes of the ocean, whoa!
Although amazingly, the majority of editing sites are non-coding (hmm)

Even in coral and octopus in response to temperature changes of the ocean, whoa!
Although amazingly, the majority of editing sites are non-coding (hmm)
There's a lot here but it's fascinating.
A to I editing is an under appreciated area of biology, where ADAR enzymes deaminate adenosine to inosine. Thousands of RNA molecules are edited all the time!

There's a lot here but it's fascinating.
A to I editing is an under appreciated area of biology, where ADAR enzymes deaminate adenosine to inosine. Thousands of RNA molecules are edited all the time!