We're looking into it :) the differences in UTR dependencies between in vitro and in cells is interesting. Big fan of your work!
April 10, 2025 at 1:13 PM
We're looking into it :) the differences in UTR dependencies between in vitro and in cells is interesting. Big fan of your work!
Lastly, I’m very thankful of my excellent co-first authors Lukas Villiger, Justin Lim, Masa Hiraizumi, and mentors Omar and Jonathan. We are so grateful for our fantastic ongoing collaborations with Hiroshi Nishimasu. Thanks to all co-authors for their invaluable contributions!
April 9, 2025 at 5:15 PM
Lastly, I’m very thankful of my excellent co-first authors Lukas Villiger, Justin Lim, Masa Hiraizumi, and mentors Omar and Jonathan. We are so grateful for our fantastic ongoing collaborations with Hiroshi Nishimasu. Thanks to all co-authors for their invaluable contributions!
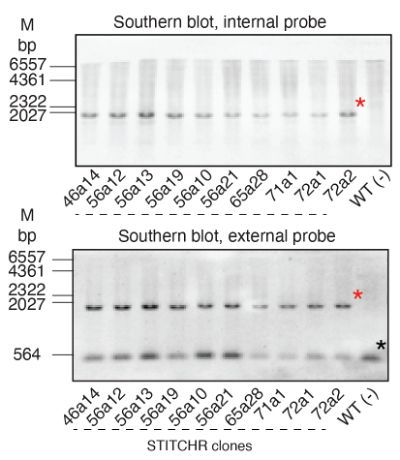
This was a really fun project which I’m excited is finally out. There were some unique challenges on the way, including learning how to Southern blot to address a reviewer’s concern. The blot got buried in the supplemental, but it took me months, so here it is in all its glory
April 9, 2025 at 5:14 PM
This was a really fun project which I’m excited is finally out. There were some unique challenges on the way, including learning how to Southern blot to address a reviewer’s concern. The blot got buried in the supplemental, but it took me months, so here it is in all its glory
There’s much more data in the paper, so please check it out! We also showed insertion by delivering RNA templates, insertion in different cell types, in vitro characterization of STITCHR, mapping of nicking sites, PacBio sequencing of long insertions, offtarget mapping and more
April 9, 2025 at 5:14 PM
There’s much more data in the paper, so please check it out! We also showed insertion by delivering RNA templates, insertion in different cell types, in vitro characterization of STITCHR, mapping of nicking sites, PacBio sequencing of long insertions, offtarget mapping and more
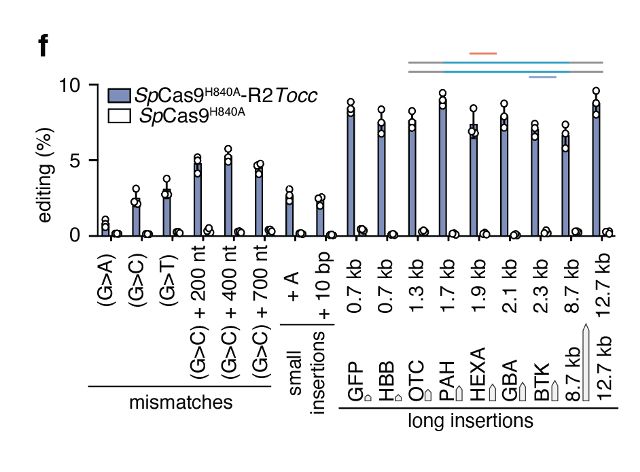
STITCHR can insert whatever is between its homology arms. We can efficiently install a diverse range of edits, including substitutions, deletions and large insertions of at least 12.7kb with high fidelity
April 9, 2025 at 5:14 PM
STITCHR can insert whatever is between its homology arms. We can efficiently install a diverse range of edits, including substitutions, deletions and large insertions of at least 12.7kb with high fidelity
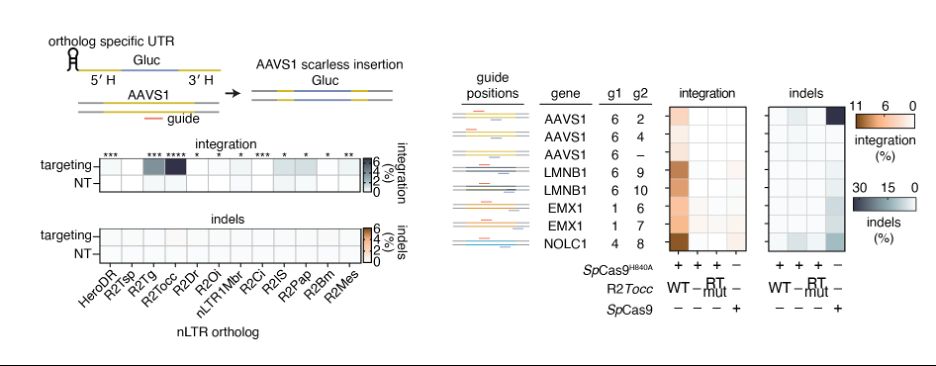
We tested a panel of R2s for their ability to perform scarless insertion at AAVS1, identifying R2Tocc as our best orthologue due to high on-target insertion and low 28S insertion. After screening sgRNA panels and homology arms, we could insert efficiently at multiple genomic loci
April 9, 2025 at 5:14 PM
We tested a panel of R2s for their ability to perform scarless insertion at AAVS1, identifying R2Tocc as our best orthologue due to high on-target insertion and low 28S insertion. After screening sgRNA panels and homology arms, we could insert efficiently at multiple genomic loci
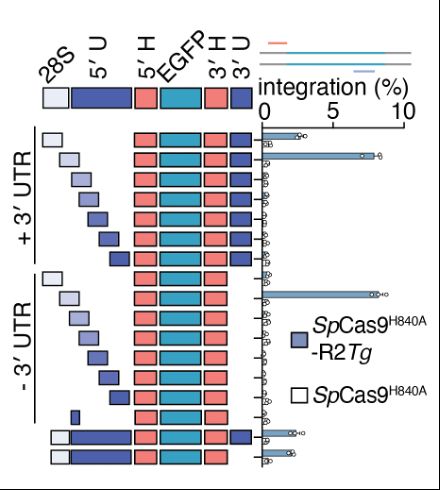
We found the architecture of the RNA cargo to be critical. The 3’ UTR is dispensable in cells, however, the homology arms and a small section of the 5’ UTR are essential, including a short sequence homologous to its natural 28S insertion site.
April 9, 2025 at 5:13 PM
We found the architecture of the RNA cargo to be critical. The 3’ UTR is dispensable in cells, however, the homology arms and a small section of the 5’ UTR are essential, including a short sequence homologous to its natural 28S insertion site.
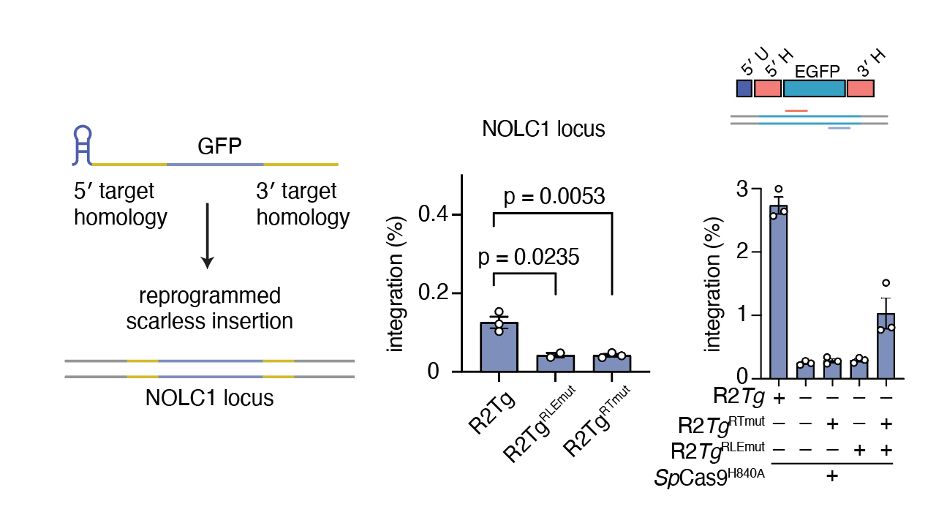
Swapping out homology arms for NOLC1 sequences led to the natural reprogramming of R2Tg, inserting a GFP at NOLC1, which was improved with nCas9-assited retargeting. We called this system Site-specific Target-primed Insertion via Targeted CRISPR Homing of Retroelements (STITCHR).
April 9, 2025 at 5:13 PM
Swapping out homology arms for NOLC1 sequences led to the natural reprogramming of R2Tg, inserting a GFP at NOLC1, which was improved with nCas9-assited retargeting. We called this system Site-specific Target-primed Insertion via Targeted CRISPR Homing of Retroelements (STITCHR).
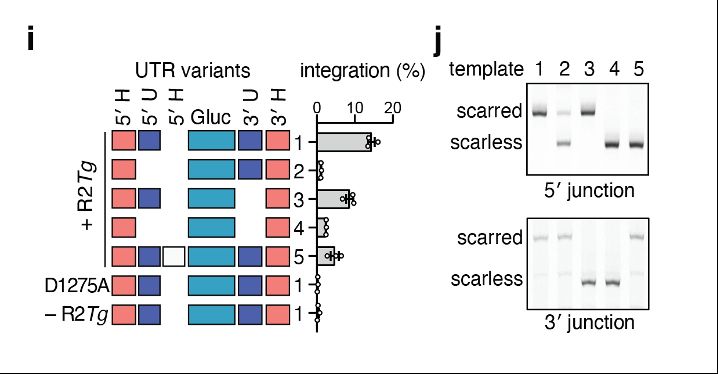
To understand cargo constraints, we permuted R2Tg’s RNA template, deleting and/or re-ordering the UTR and homology arm architecture. Interestingly, deletion of the UTRs internal to the homology arms were still functional and resulted in scarless cargo insertion!
April 9, 2025 at 5:13 PM
To understand cargo constraints, we permuted R2Tg’s RNA template, deleting and/or re-ordering the UTR and homology arm architecture. Interestingly, deletion of the UTRs internal to the homology arms were still functional and resulted in scarless cargo insertion!
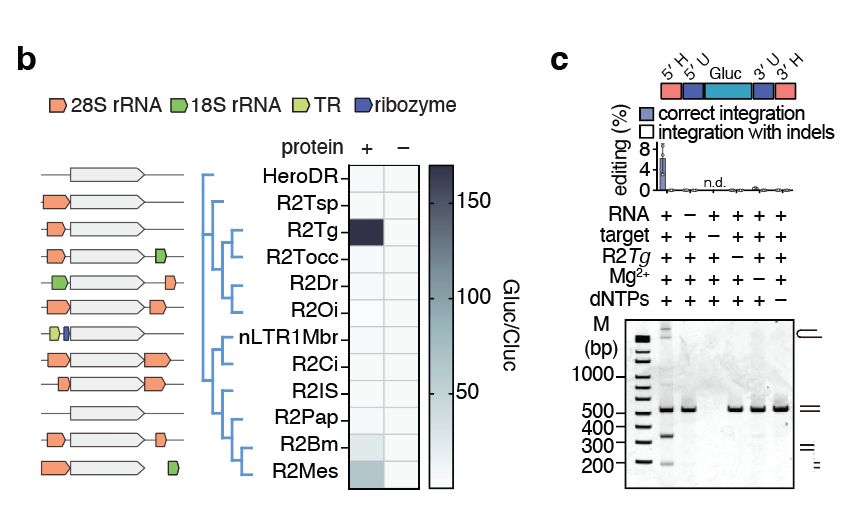
We selected a diverse set of R2s and tested if they could insert non-natural cargoes at their natural sites in human cells. We identified an R2 from the zebra finch Taeniopygia guttata (R2Tg) with high insertion activity in cells and in vitro.
April 9, 2025 at 5:13 PM
We selected a diverse set of R2s and tested if they could insert non-natural cargoes at their natural sites in human cells. We identified an R2 from the zebra finch Taeniopygia guttata (R2Tg) with high insertion activity in cells and in vitro.
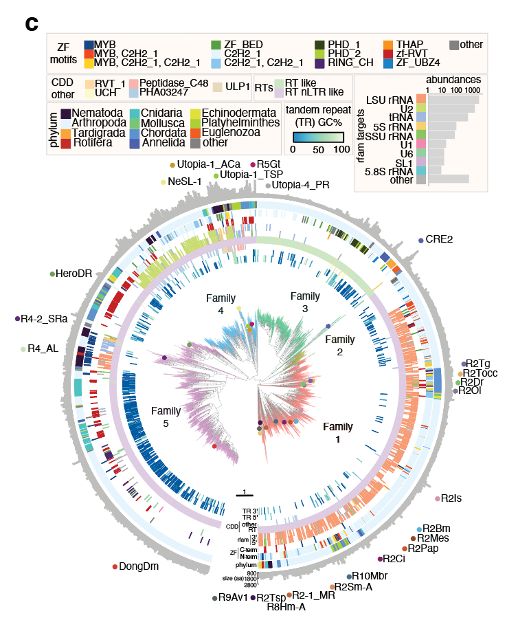
We were interested in whether R2s could be reprogrammed in cells. We searched the natural world, uncovering 8,248 orthologues and their site preferences. Excitingly, we discovered instances of R2s acquiring novel insertion sites during evolution, a natural form of reprogramming!
April 9, 2025 at 5:12 PM
We were interested in whether R2s could be reprogrammed in cells. We searched the natural world, uncovering 8,248 orthologues and their site preferences. Excitingly, we discovered instances of R2s acquiring novel insertion sites during evolution, a natural form of reprogramming!
Additionally, fascinating work from the Zhang lab reported the structure of Bombyx mori R2 and showed that in principle R2s could be reprogrammed, at least in vitro.
www.science.org/doi/10.1126/...
www.science.org/doi/10.1126/...
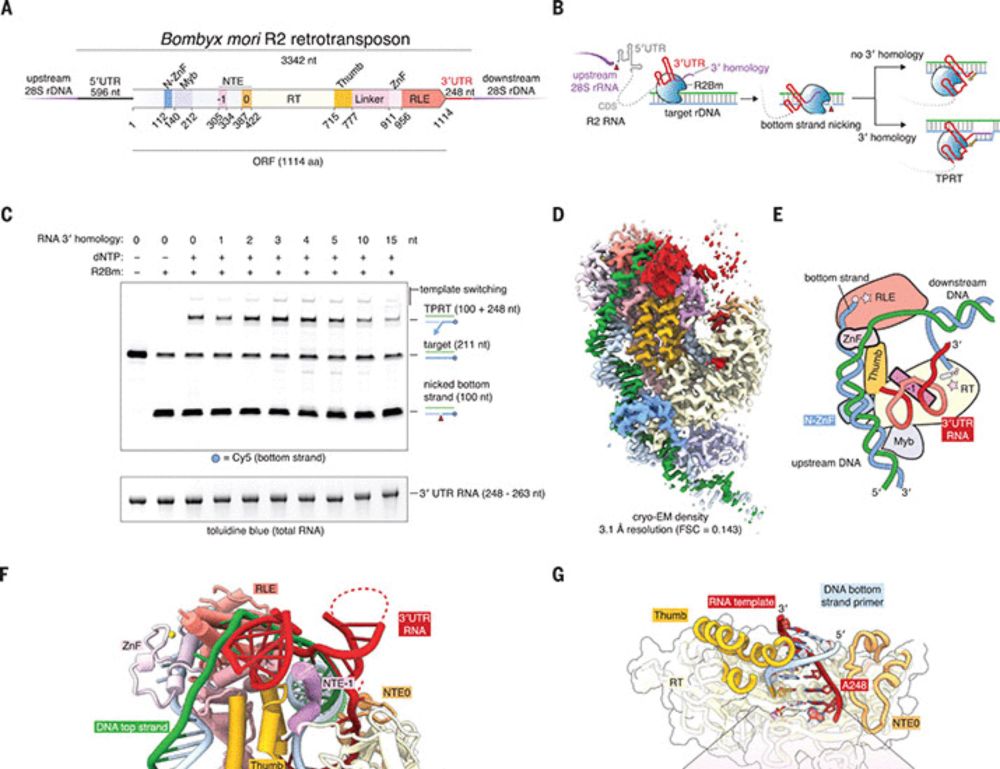
Structure of the R2 non-LTR retrotransposon initiating target-primed reverse transcription
A retrotransposon structure elucidates the principles of target DNA selection and self-RNA recognition.
www.science.org
April 9, 2025 at 5:12 PM
Additionally, fascinating work from the Zhang lab reported the structure of Bombyx mori R2 and showed that in principle R2s could be reprogrammed, at least in vitro.
www.science.org/doi/10.1126/...
www.science.org/doi/10.1126/...
Impressive work from the Collins lab and others demonstrated the repurposing of R2s for all-RNA insertion of transgenes at human safe harbour loci, an exciting therapeutic application!
www.nature.com/articles/s41...
www.nature.com/articles/s41...
Harnessing eukaryotic retroelement proteins for transgene insertion into human safe-harbor loci - Nature Biotechnology
Transgenes are inserted into human cells by 2-RNA delivery of a retroelement protein and template.
www.nature.com
April 9, 2025 at 5:12 PM
Impressive work from the Collins lab and others demonstrated the repurposing of R2s for all-RNA insertion of transgenes at human safe harbour loci, an exciting therapeutic application!
www.nature.com/articles/s41...
www.nature.com/articles/s41...
In the AbuGoot lab, we were intrigued by the R2 retrotransposon class of mobile genetic elements. Foundational work by Thomas Eikbush and colleagues showed a Target-Primed Reverse Transcription mechanism of insertion, priming with RNA homology arms.
April 9, 2025 at 5:11 PM
In the AbuGoot lab, we were intrigued by the R2 retrotransposon class of mobile genetic elements. Foundational work by Thomas Eikbush and colleagues showed a Target-Primed Reverse Transcription mechanism of insertion, priming with RNA homology arms.

