
❤️ ubiquitin, chaperones & proteomics
💙 outdoor, repast, electro-rock & traveling 🇨🇭🇨🇦
https://mayor51.wixsite.com/mayorlab
Happy birthday- enjoy the rest of the trip
Happy birthday- enjoy the rest of the trip
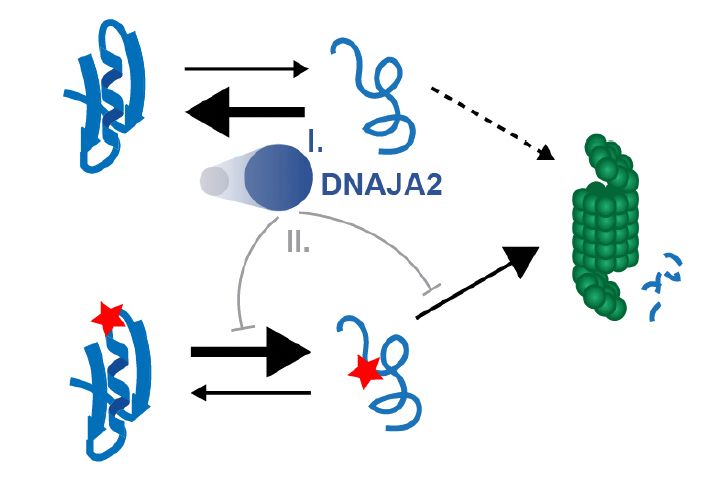
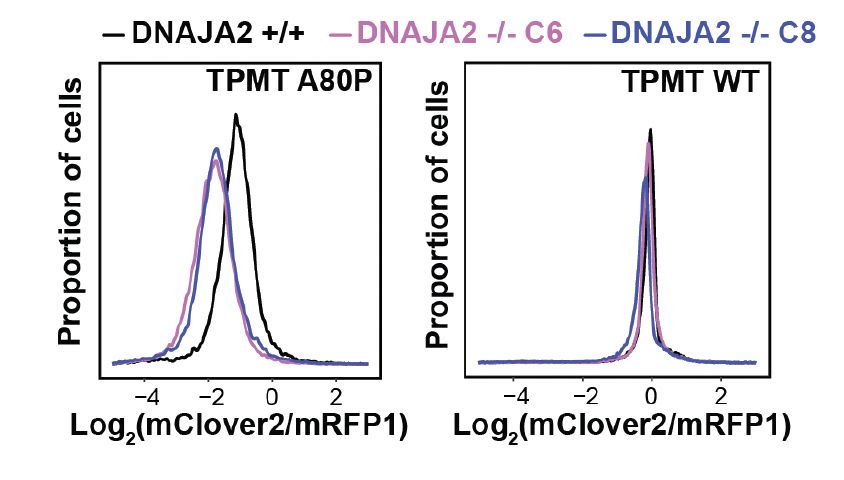
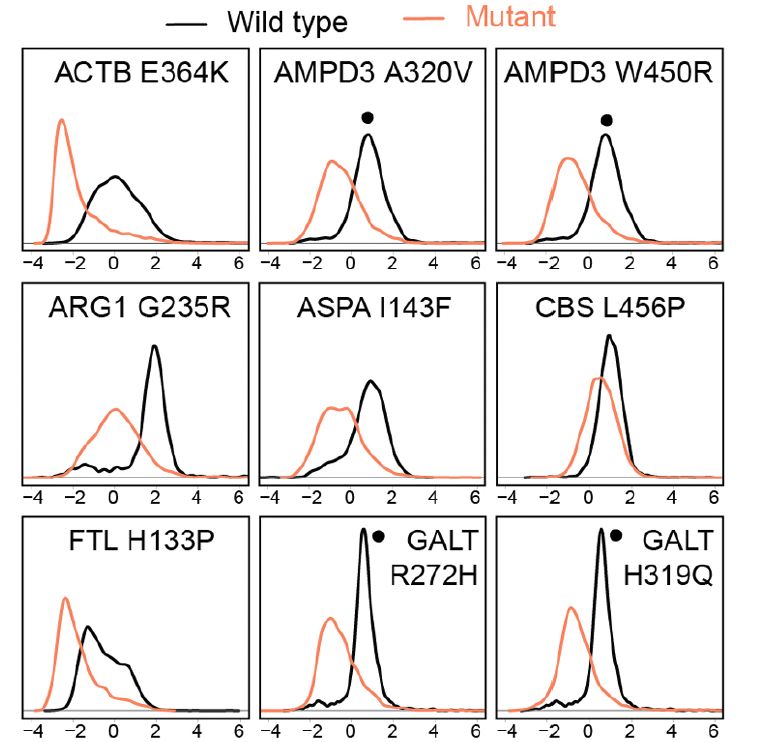
The illustration was created by @taniodraws.bsky.social our go-to freelance artist for summarizing our studies.
The illustration was created by @taniodraws.bsky.social our go-to freelance artist for summarizing our studies.
We first need to better educate our trainees about the issue (e.g. does your grad program cover the topic in a course?)
We first need to better educate our trainees about the issue (e.g. does your grad program cover the topic in a course?)

