with Matthias Feige and Brenda Schulman
Proteostasis and membrane protein enthusiast
Boehringer Ingelheim Fonds PhD fellow

ATP binding stabilizes Spf1’s “arm” domain, contacting EMC’s cytoplasmic cap above the insertase cavity, closing the composite cavity.

ATP binding stabilizes Spf1’s “arm” domain, contacting EMC’s cytoplasmic cap above the insertase cavity, closing the composite cavity.




#ERliterature #chaperone #proteostasis
9/9
#ERliterature #chaperone #proteostasis
9/9
We found that challenging TMDs remain bound to EMC and are ER-retained—but once a partner for productive assembly is available, EMC binding is reduced and the protein can exit the ER.
8/9
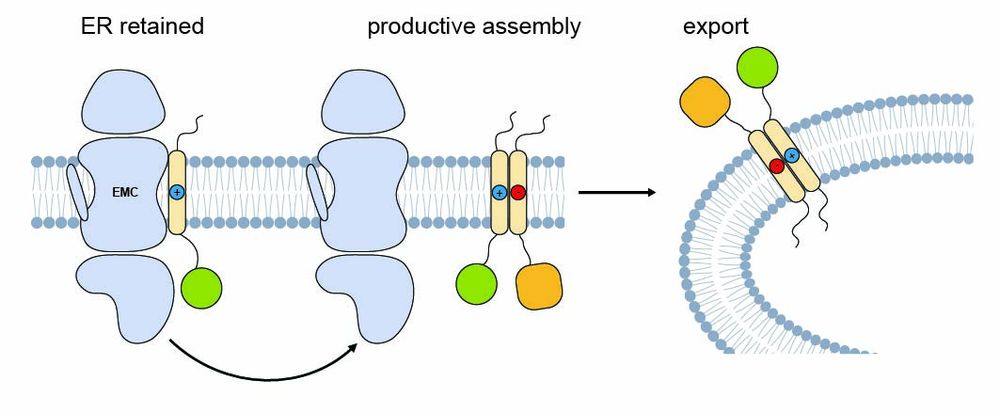
We found that challenging TMDs remain bound to EMC and are ER-retained—but once a partner for productive assembly is available, EMC binding is reduced and the protein can exit the ER.
8/9
7/9
7/9
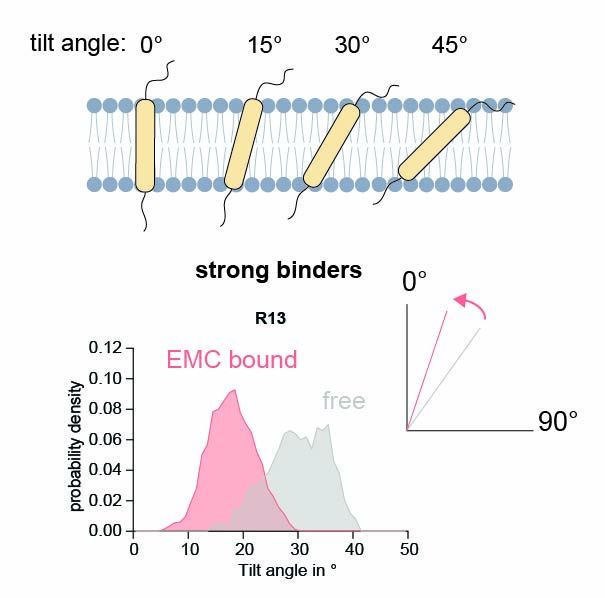
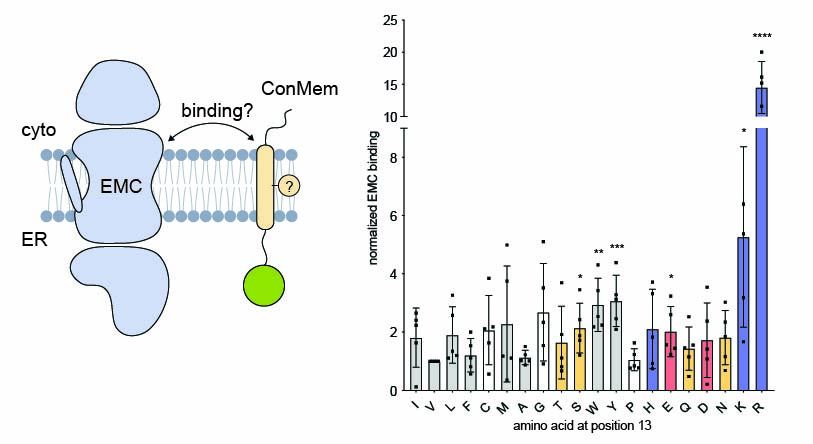
Surprisingly, mutational analysis and site-specific crosslinking showed that EMC doesn't bind the polar face of the TMD—but engages the opposite, hydrophobic side.
6/9
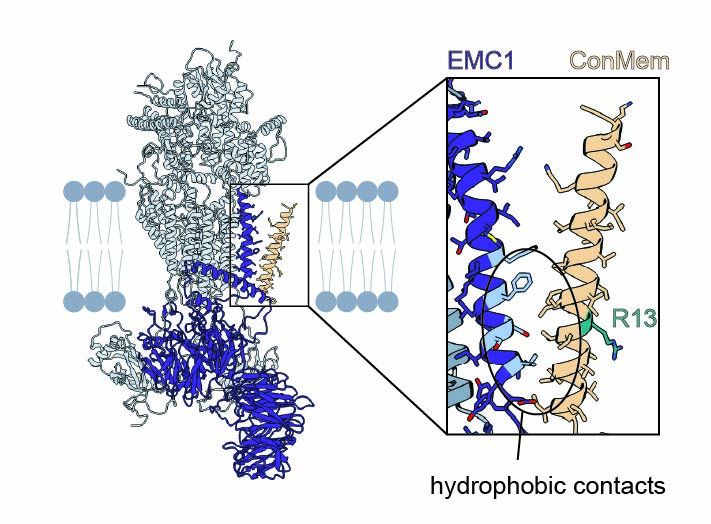
Surprisingly, mutational analysis and site-specific crosslinking showed that EMC doesn't bind the polar face of the TMD—but engages the opposite, hydrophobic side.
6/9
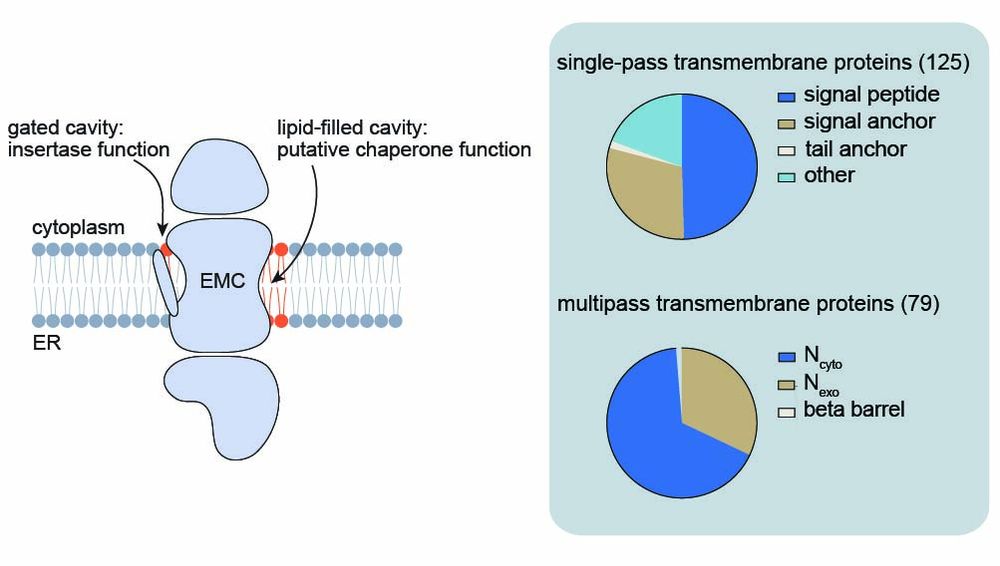
Their TMDs often contain polar/charged residues needed for function but are only marginally stable in the membrane—making them ideal candidates for chaperone support during folding and assembly.
5/9
Their TMDs often contain polar/charged residues needed for function but are only marginally stable in the membrane—making them ideal candidates for chaperone support during folding and assembly.
5/9
We trained and validated a neural network (ipredEMC) to predict EMC binding proteome-wide. This tool revealed that transporters and ion channels are major chaperone clients.
4/9
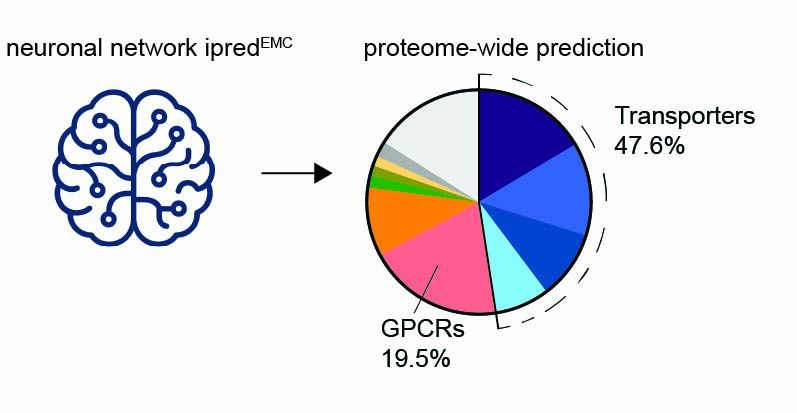
We trained and validated a neural network (ipredEMC) to predict EMC binding proteome-wide. This tool revealed that transporters and ion channels are major chaperone clients.
4/9
3/9
3/9
2/9
2/9

