
Jesse Shapiro
@bjesseshapiro.bsky.social
If you’re at @astmh.bsky.social #ASTMH2025 check out our session today on Enteric Pathogen Genomics & Evolution (room 717, 16:30) featuring @ksbakes.bsky.social on Shigella, plus 2 other great talks on V. cholerae phages, and Salmonella!

November 10, 2025 at 2:04 PM
If you’re at @astmh.bsky.social #ASTMH2025 check out our session today on Enteric Pathogen Genomics & Evolution (room 717, 16:30) featuring @ksbakes.bsky.social on Shigella, plus 2 other great talks on V. cholerae phages, and Salmonella!
Great view of the first snow of the season on the train from Montreal to Toronto for @astmh.bsky.social #ASTMH2025. Maybe in my lifetime this trip will take 3 hours instead of 5…
November 9, 2025 at 7:09 PM
Great view of the first snow of the season on the train from Montreal to Toronto for @astmh.bsky.social #ASTMH2025. Maybe in my lifetime this trip will take 3 hours instead of 5…
We heard the Kassen lab’s sequencer was being badly neglected so we have adopted it.
It’s being fed a healthy diet of high molecular weight DNA.
Will return it when your computer is fixed @reeskassen.bsky.social ;)
It’s being fed a healthy diet of high molecular weight DNA.
Will return it when your computer is fixed @reeskassen.bsky.social ;)

November 7, 2025 at 10:31 PM
We heard the Kassen lab’s sequencer was being badly neglected so we have adopted it.
It’s being fed a healthy diet of high molecular weight DNA.
Will return it when your computer is fixed @reeskassen.bsky.social ;)
It’s being fed a healthy diet of high molecular weight DNA.
Will return it when your computer is fixed @reeskassen.bsky.social ;)
circular AI shrimp are decidedly cursed

October 26, 2025 at 5:11 PM
circular AI shrimp are decidedly cursed
Sabbatical planning: step 1.

October 7, 2025 at 8:24 PM
Sabbatical planning: step 1.

September 25, 2025 at 11:46 PM
The New Vic project @mcgill.ca is entering an exciting, noisy and rather smashy phase. Cool to watch but I’m glad my lab isn’t right across the street…

September 5, 2025 at 12:54 AM
The New Vic project @mcgill.ca is entering an exciting, noisy and rather smashy phase. Cool to watch but I’m glad my lab isn’t right across the street…
I prefer to avoid driving when possible. But when I do, I like the car to look like mostly bikes.

September 3, 2025 at 1:28 AM
I prefer to avoid driving when possible. But when I do, I like the car to look like mostly bikes.
Tying this all together in regression model, we see that co-occurrence is the strongest driver of HGT even after controlling for phylogeny (which also has a big effect). Co-occurrence is the main driver even after controlling for measured environmental factors
19/n
19/n

August 21, 2025 at 5:59 PM
Tying this all together in regression model, we see that co-occurrence is the strongest driver of HGT even after controlling for phylogeny (which also has a big effect). Co-occurrence is the main driver even after controlling for measured environmental factors
19/n
19/n
An HGT event might be adaptive under specific environmental conditions, making it more likely to be retained and observed. Indeed, genome pairs connected by HGT tend to be sampled under more similar environmental conditions compared to genomes without evidence for HGT.
18/n
18/n

August 21, 2025 at 5:57 PM
An HGT event might be adaptive under specific environmental conditions, making it more likely to be retained and observed. Indeed, genome pairs connected by HGT tend to be sampled under more similar environmental conditions compared to genomes without evidence for HGT.
18/n
18/n
Using >15k whole-genome sequences and >1,800 metagenomes from oceans, we can see the effect of phylogeny, with more HGT events found between closer relatives.
16/n
16/n

August 21, 2025 at 5:52 PM
Using >15k whole-genome sequences and >1,800 metagenomes from oceans, we can see the effect of phylogeny, with more HGT events found between closer relatives.
16/n
16/n
Summary of parts 1 & @ (gene-centric perspective): even rare genes in the pangenome can be adaptive to the host (we estimate 2-20% of genes; see the paper for details!) and there is a lot of variation across gene functions!
12/n
12/n

August 21, 2025 at 5:40 PM
Summary of parts 1 & @ (gene-centric perspective): even rare genes in the pangenome can be adaptive to the host (we estimate 2-20% of genes; see the paper for details!) and there is a lot of variation across gene functions!
12/n
12/n
Mobilome (X) is strongly enriched in pseudogenes, but most of this effect is due to transposons, not phage-related genes, being pseudogenized. This is consistent with phages often encoding genes that are adaptive to their host genome, as suggested previously:
academic.oup.com/mbe/article/...
11/n
academic.oup.com/mbe/article/...
11/n

August 21, 2025 at 5:38 PM
Mobilome (X) is strongly enriched in pseudogenes, but most of this effect is due to transposons, not phage-related genes, being pseudogenized. This is consistent with phages often encoding genes that are adaptive to their host genome, as suggested previously:
academic.oup.com/mbe/article/...
11/n
academic.oup.com/mbe/article/...
11/n
Key finding: Nearly all COG categories are depleted in pseudogenes when there is no redundant copy of their gene family.
Interpretation: Even very rare accessory genes are under selection to maintain a functional copy in the genome.
10/n
Interpretation: Even very rare accessory genes are under selection to maintain a functional copy in the genome.
10/n

August 21, 2025 at 5:34 PM
Key finding: Nearly all COG categories are depleted in pseudogenes when there is no redundant copy of their gene family.
Interpretation: Even very rare accessory genes are under selection to maintain a functional copy in the genome.
10/n
Interpretation: Even very rare accessory genes are under selection to maintain a functional copy in the genome.
10/n
The interpretation? Mobilome genes likely spread rapidly across species barriers. Defence and secondary metabolism genes may be enriched in intermediate-frequency mutations as they spread across species, maybe due to balancing selection or species-specific selective pressures.
7/n
7/n

August 21, 2025 at 5:25 PM
The interpretation? Mobilome genes likely spread rapidly across species barriers. Defence and secondary metabolism genes may be enriched in intermediate-frequency mutations as they spread across species, maybe due to balancing selection or species-specific selective pressures.
7/n
7/n
WAIT BUT in the real data there is a lot of variation across functional gene categories: most (shades of red, like category X: mobilome) show a negative correlation between mobility and Tajima's D, but others show a positive correlation (shades of blue, like V: defence and Q: secondary metab).
6/n
6/n

August 21, 2025 at 5:21 PM
WAIT BUT in the real data there is a lot of variation across functional gene categories: most (shades of red, like category X: mobilome) show a negative correlation between mobility and Tajima's D, but others show a positive correlation (shades of blue, like V: defence and Q: secondary metab).
6/n
6/n
In general, gene mobility is negatively correlated with Tajima's D -- suggesting that mobile genes spread rapidly* across many species in the microbiome, presumably due to positive selection
*before many mutations can reach high frequency
(measured using Tajima’s D)
4/n
*before many mutations can reach high frequency
(measured using Tajima’s D)
4/n

August 21, 2025 at 5:13 PM
In general, gene mobility is negatively correlated with Tajima's D -- suggesting that mobile genes spread rapidly* across many species in the microbiome, presumably due to positive selection
*before many mutations can reach high frequency
(measured using Tajima’s D)
4/n
*before many mutations can reach high frequency
(measured using Tajima’s D)
4/n
Let's start by thinking of levels of selection: from an individual gene (gene-centric) to an entire pangenome (species-centric). These perspectives are explored in this 2021 piece with @gmdouglas.bsky.social -- also the lead author of parts 2 & 3 of this talk!
doi.org/10.1093/gbe/...
2/n
doi.org/10.1093/gbe/...
2/n

August 21, 2025 at 5:04 PM
Let's start by thinking of levels of selection: from an individual gene (gene-centric) to an entire pangenome (species-centric). These perspectives are explored in this 2021 piece with @gmdouglas.bsky.social -- also the lead author of parts 2 & 3 of this talk!
doi.org/10.1093/gbe/...
2/n
doi.org/10.1093/gbe/...
2/n
Notably, two AMR genes on the ICE (qnrVc and mphA) that were previously associated with ciprofloxacin and azithromycin resistance in a 2006 sampling of V. cholerae genomes from Dhaka were absent in the 2018–19 genomes.
6/n
6/n

July 25, 2025 at 2:36 PM
Notably, two AMR genes on the ICE (qnrVc and mphA) that were previously associated with ciprofloxacin and azithromycin resistance in a 2006 sampling of V. cholerae genomes from Dhaka were absent in the 2018–19 genomes.
6/n
6/n
BD3 is distantly related (phylogenetically) with BD1, but encodes a similar profile of antibiotic and phage resistance genes suggesting they could compete in similar niches. This contrasts with BD2 which generally encodes fewer resistance genes.
5/n
5/n

July 25, 2025 at 2:30 PM
BD3 is distantly related (phylogenetically) with BD1, but encodes a similar profile of antibiotic and phage resistance genes suggesting they could compete in similar niches. This contrasts with BD2 which generally encodes fewer resistance genes.
5/n
5/n
BD1 is associated with more severe disease and is more prevalent in the North, consistent with an Indian origin (also supported by phylogenetic analysis). There is more genomic diversity ("genomic clusters," which are roughly equivalent to lineages but include pangenome variation) in the South.
4/n
4/n
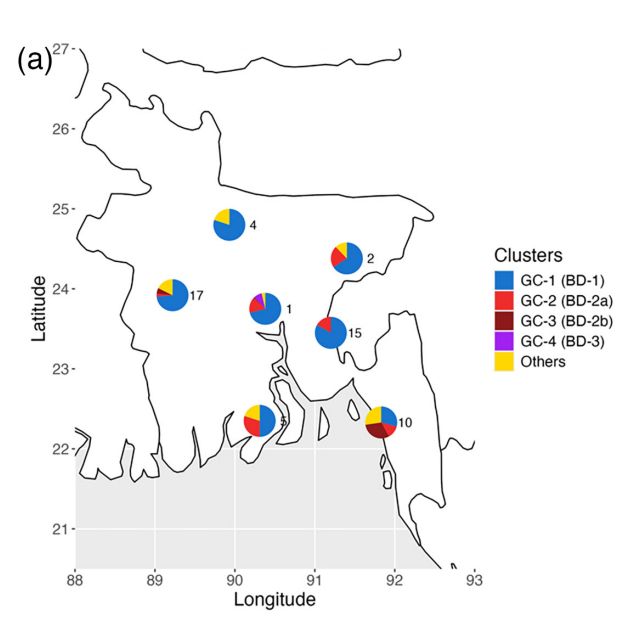
July 25, 2025 at 2:24 PM
BD1 is associated with more severe disease and is more prevalent in the North, consistent with an Indian origin (also supported by phylogenetic analysis). There is more genomic diversity ("genomic clusters," which are roughly equivalent to lineages but include pangenome variation) in the South.
4/n
4/n
Each lineage is associated with different Integrative Conjugative Element (ICE) types, with occasional gain/loss events. The lineages are widely distributed across Bangladesh, with the new lineage BD3 mostly restricted to Dhaka in 2019.
3/n
3/n

July 25, 2025 at 2:17 PM
Each lineage is associated with different Integrative Conjugative Element (ICE) types, with occasional gain/loss events. The lineages are widely distributed across Bangladesh, with the new lineage BD3 mostly restricted to Dhaka in 2019.
3/n
3/n
Can we talk about Garagecore as a style of architecture?

July 24, 2025 at 4:16 PM
Can we talk about Garagecore as a style of architecture?



