Bioinformatics Advances
@bioinfoadv.bsky.social
A fully open access, peer-reviewed journal published jointly by Oxford University Press and the International Society for Computational Biology.
🧬 Now published in Bioinformatics Advances: “LSTM-based deep learning model for the discovery of antimicrobial peptides targeting Mycobacterium tuberculosis.”
Read the full paper here: https://doi.org/10.1093/bioadv/vbaf274
Authors include: @taanec.bsky.social
Read the full paper here: https://doi.org/10.1093/bioadv/vbaf274
Authors include: @taanec.bsky.social

November 11, 2025 at 10:01 AM
🧬 Now published in Bioinformatics Advances: “LSTM-based deep learning model for the discovery of antimicrobial peptides targeting Mycobacterium tuberculosis.”
Read the full paper here: https://doi.org/10.1093/bioadv/vbaf274
Authors include: @taanec.bsky.social
Read the full paper here: https://doi.org/10.1093/bioadv/vbaf274
Authors include: @taanec.bsky.social
🧠 Just out in Bioinformatics Advances: “Deep learning for regulatory genomics: A survey of models, challenges, and applications.”
Explore the full study: https://doi.org/10.1093/bioadv/vbaf271
Explore the full study: https://doi.org/10.1093/bioadv/vbaf271

November 10, 2025 at 10:02 AM
🧠 Just out in Bioinformatics Advances: “Deep learning for regulatory genomics: A survey of models, challenges, and applications.”
Explore the full study: https://doi.org/10.1093/bioadv/vbaf271
Explore the full study: https://doi.org/10.1093/bioadv/vbaf271
🧩 Explore the latest from Bioinformatics Advances: “CoRTE: A web-service for constructing temporal networks from genotype-tissue expression data.”
Full article available: https://doi.org/10.1093/bioadv/vbaf272
Full article available: https://doi.org/10.1093/bioadv/vbaf272

November 7, 2025 at 10:02 AM
🧩 Explore the latest from Bioinformatics Advances: “CoRTE: A web-service for constructing temporal networks from genotype-tissue expression data.”
Full article available: https://doi.org/10.1093/bioadv/vbaf272
Full article available: https://doi.org/10.1093/bioadv/vbaf272
🧠 Now published in Bioinformatics Advances: “PSO-FeatureFusion: A general framework for fusing heterogeneous features via particle swarm optimization.”
Read the full paper here: https://doi.org/10.1093/bioadv/vbaf263
Read the full paper here: https://doi.org/10.1093/bioadv/vbaf263

November 6, 2025 at 10:02 AM
🧠 Now published in Bioinformatics Advances: “PSO-FeatureFusion: A general framework for fusing heterogeneous features via particle swarm optimization.”
Read the full paper here: https://doi.org/10.1093/bioadv/vbaf263
Read the full paper here: https://doi.org/10.1093/bioadv/vbaf263
🔥 Now published in Bioinformatics Advances: “Shiny-Calorie: A context-aware application for indirect calorimetry data analysis and visualization using R.”
Read the full paper here: https://doi.org/10.1093/bioadv/vbaf270
Authors include: @wachtenlab.bsky.social, @ecsignalmetabolism.bsky.social
Read the full paper here: https://doi.org/10.1093/bioadv/vbaf270
Authors include: @wachtenlab.bsky.social, @ecsignalmetabolism.bsky.social

November 5, 2025 at 10:01 AM
🔥 Now published in Bioinformatics Advances: “Shiny-Calorie: A context-aware application for indirect calorimetry data analysis and visualization using R.”
Read the full paper here: https://doi.org/10.1093/bioadv/vbaf270
Authors include: @wachtenlab.bsky.social, @ecsignalmetabolism.bsky.social
Read the full paper here: https://doi.org/10.1093/bioadv/vbaf270
Authors include: @wachtenlab.bsky.social, @ecsignalmetabolism.bsky.social
💊 Now published in Bioinformatics Advances: “DSA-DeepFM: A dual-stage attention-enhanced DeepFM model for predicting anticancer synergistic drug combinations.”
Read the full paper here: https://doi.org/10.1093/bioadv/vbaf269
Read the full paper here: https://doi.org/10.1093/bioadv/vbaf269

November 4, 2025 at 10:02 AM
💊 Now published in Bioinformatics Advances: “DSA-DeepFM: A dual-stage attention-enhanced DeepFM model for predicting anticancer synergistic drug combinations.”
Read the full paper here: https://doi.org/10.1093/bioadv/vbaf269
Read the full paper here: https://doi.org/10.1093/bioadv/vbaf269
🧬 Just out in Bioinformatics Advances: “TidyGWAS: A scalable approach for standardized cleaning of genome-wide association study summary statistics.”
Explore the full study: https://doi.org/10.1093/bioadv/vbaf262
Authors include: @joellepasman.bsky.social
Explore the full study: https://doi.org/10.1093/bioadv/vbaf262
Authors include: @joellepasman.bsky.social

November 3, 2025 at 10:01 AM
🧬 Just out in Bioinformatics Advances: “TidyGWAS: A scalable approach for standardized cleaning of genome-wide association study summary statistics.”
Explore the full study: https://doi.org/10.1093/bioadv/vbaf262
Authors include: @joellepasman.bsky.social
Explore the full study: https://doi.org/10.1093/bioadv/vbaf262
Authors include: @joellepasman.bsky.social
🧠 Now published in Bioinformatics Advances: “Gaining insights into Alzheimer’s disease by predicting chromatin spatial organization.”
Read the full paper here: ttps://doi.org/10.1093/bioadv/vbaf268
Read the full paper here: ttps://doi.org/10.1093/bioadv/vbaf268

October 31, 2025 at 10:02 AM
🧠 Now published in Bioinformatics Advances: “Gaining insights into Alzheimer’s disease by predicting chromatin spatial organization.”
Read the full paper here: ttps://doi.org/10.1093/bioadv/vbaf268
Read the full paper here: ttps://doi.org/10.1093/bioadv/vbaf268
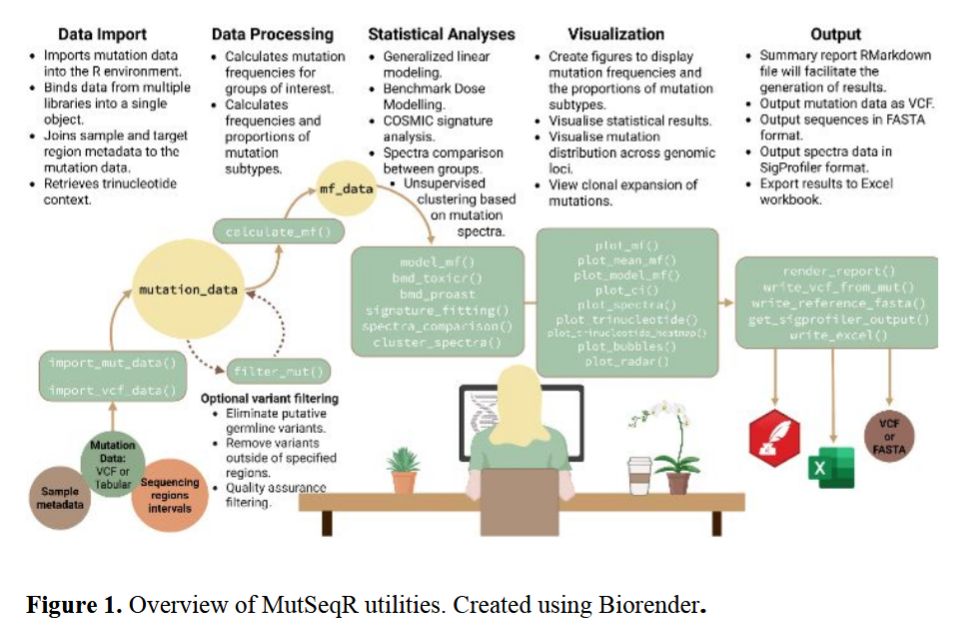
🧬 Explore the latest from Bioinformatics Advances: “MutSeqR: An open-source R package for standardized analysis of error-corrected next-generation sequencing data in genetic toxicology.”
Full article available: https://doi.org/10.1093/bioadv/vbaf265
Full article available: https://doi.org/10.1093/bioadv/vbaf265
October 31, 2025 at 9:02 AM
🧬 Explore the latest from Bioinformatics Advances: “MutSeqR: An open-source R package for standardized analysis of error-corrected next-generation sequencing data in genetic toxicology.”
Full article available: https://doi.org/10.1093/bioadv/vbaf265
Full article available: https://doi.org/10.1093/bioadv/vbaf265
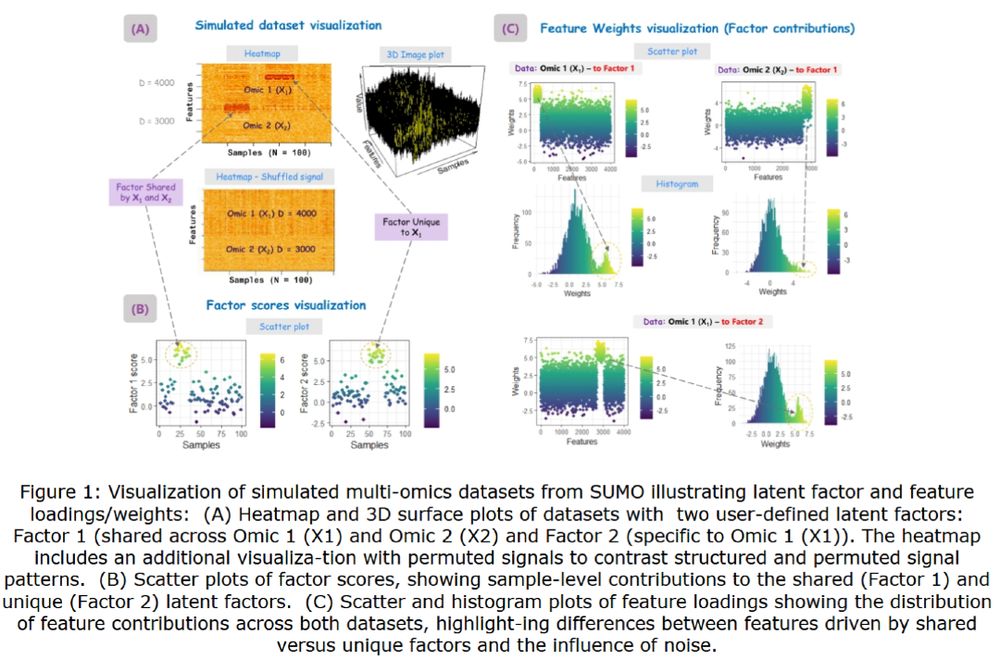
🧪 Now published in Bioinformatics Advances: “SUMO: An R package for simulating multi-omics data for methods development and testing.”
Read the full paper here: https://doi.org/10.1093/bioadv/vbaf264
Read the full paper here: https://doi.org/10.1093/bioadv/vbaf264
October 30, 2025 at 10:02 AM
🧪 Now published in Bioinformatics Advances: “SUMO: An R package for simulating multi-omics data for methods development and testing.”
Read the full paper here: https://doi.org/10.1093/bioadv/vbaf264
Read the full paper here: https://doi.org/10.1093/bioadv/vbaf264
🧬 Just out in Bioinformatics Advances: “Unifying proteomic technologies with ProteinProjector.”
Explore the full study: https://doi.org/10.1093/bioadv/vbaf266
Explore the full study: https://doi.org/10.1093/bioadv/vbaf266

October 30, 2025 at 9:02 AM
🧬 Just out in Bioinformatics Advances: “Unifying proteomic technologies with ProteinProjector.”
Explore the full study: https://doi.org/10.1093/bioadv/vbaf266
Explore the full study: https://doi.org/10.1093/bioadv/vbaf266
🧩 Explore the latest from Bioinformatics Advances: “StarPepWeb: An integrative, graph-based resource for bioactive peptides.”
Full article available: https://doi.org/10.1093/bioadv/vbaf261
Full article available: https://doi.org/10.1093/bioadv/vbaf261

October 29, 2025 at 10:01 AM
🧩 Explore the latest from Bioinformatics Advances: “StarPepWeb: An integrative, graph-based resource for bioactive peptides.”
Full article available: https://doi.org/10.1093/bioadv/vbaf261
Full article available: https://doi.org/10.1093/bioadv/vbaf261
🌾 Now published in Bioinformatics Advances: “NifFinder: Improved Nif protein prediction using SWeeP vectors and neural networks.”
Read the full paper here: https://doi.org/10.1093/bioadv/vbaf260
Read the full paper here: https://doi.org/10.1093/bioadv/vbaf260

October 29, 2025 at 9:02 AM
🌾 Now published in Bioinformatics Advances: “NifFinder: Improved Nif protein prediction using SWeeP vectors and neural networks.”
Read the full paper here: https://doi.org/10.1093/bioadv/vbaf260
Read the full paper here: https://doi.org/10.1093/bioadv/vbaf260
🌊 Now published in Bioinformatics Advances: “The Hydractinia Genome Project Portal: Multi-omic annotation and visualization of Hydractinia genomic datasets.”
Read the full paper here: https://doi.org/10.1093/bioadv/vbaf215
Read the full paper here: https://doi.org/10.1093/bioadv/vbaf215

October 28, 2025 at 10:01 AM
🌊 Now published in Bioinformatics Advances: “The Hydractinia Genome Project Portal: Multi-omic annotation and visualization of Hydractinia genomic datasets.”
Read the full paper here: https://doi.org/10.1093/bioadv/vbaf215
Read the full paper here: https://doi.org/10.1093/bioadv/vbaf215
🧠 Explore the latest from Bioinformatics Advances: “ICCTax: A hierarchical taxonomic classifier for metagenomic sequences on a large language model.”
Full article available: https://doi.org/10.1093/bioadv/vbaf257
Full article available: https://doi.org/10.1093/bioadv/vbaf257

October 28, 2025 at 9:03 AM
🧠 Explore the latest from Bioinformatics Advances: “ICCTax: A hierarchical taxonomic classifier for metagenomic sequences on a large language model.”
Full article available: https://doi.org/10.1093/bioadv/vbaf257
Full article available: https://doi.org/10.1093/bioadv/vbaf257
🪶 Just out in Bioinformatics Advances: “In silico analysis of insect-associated bacterial phytases reveals optimal biochemical properties and function in poultry gut condition.”
Explore the full study: https://doi.org/10.1093/bioadv/vbaf256
Explore the full study: https://doi.org/10.1093/bioadv/vbaf256

October 27, 2025 at 10:01 AM
🪶 Just out in Bioinformatics Advances: “In silico analysis of insect-associated bacterial phytases reveals optimal biochemical properties and function in poultry gut condition.”
Explore the full study: https://doi.org/10.1093/bioadv/vbaf256
Explore the full study: https://doi.org/10.1093/bioadv/vbaf256
🧪 Now published in Bioinformatics Advances: “Pathogenicity patterns in cytochrome P450 family.”
Read the full paper here: https://doi.org/10.1093/bioadv/vbaf231
Authors include: @krapnik.bsky.social
Read the full paper here: https://doi.org/10.1093/bioadv/vbaf231
Authors include: @krapnik.bsky.social

October 27, 2025 at 9:01 AM
🧪 Now published in Bioinformatics Advances: “Pathogenicity patterns in cytochrome P450 family.”
Read the full paper here: https://doi.org/10.1093/bioadv/vbaf231
Authors include: @krapnik.bsky.social
Read the full paper here: https://doi.org/10.1093/bioadv/vbaf231
Authors include: @krapnik.bsky.social
🧠 Explore the latest from Bioinformatics Advances: “Asymmetric integration of various cancer datasets for identifying risk-associated variants and genes.”
Full article available: https://doi.org/10.1093/bioadv/vbaf253
Full article available: https://doi.org/10.1093/bioadv/vbaf253

October 24, 2025 at 10:01 AM
🧠 Explore the latest from Bioinformatics Advances: “Asymmetric integration of various cancer datasets for identifying risk-associated variants and genes.”
Full article available: https://doi.org/10.1093/bioadv/vbaf253
Full article available: https://doi.org/10.1093/bioadv/vbaf253
🧬 Now published in Bioinformatics Advances: “ProtFun: A protein function prediction model using graph attention networks with a protein large language model.”
Read the full paper here: https://doi.org/10.1093/bioadv/vbaf245
Read the full paper here: https://doi.org/10.1093/bioadv/vbaf245

October 24, 2025 at 9:01 AM
🧬 Now published in Bioinformatics Advances: “ProtFun: A protein function prediction model using graph attention networks with a protein large language model.”
Read the full paper here: https://doi.org/10.1093/bioadv/vbaf245
Read the full paper here: https://doi.org/10.1093/bioadv/vbaf245
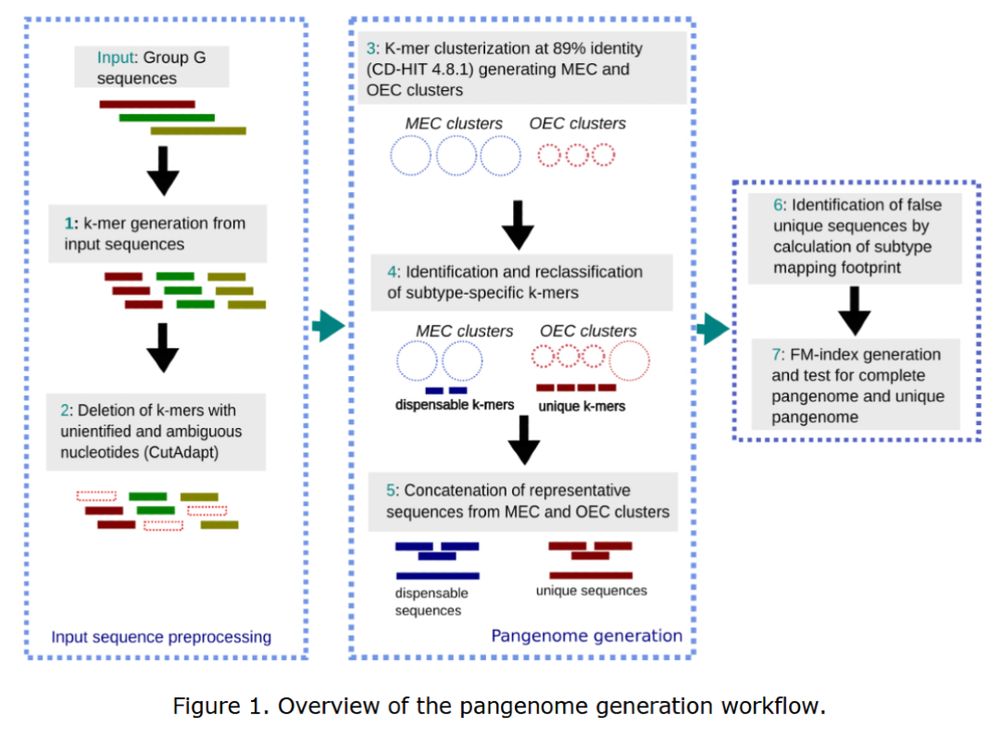
🦠 Now published in Bioinformatics Advances: “K-FluDB: A novel k-mer based database for enhanced genomic surveillance of influenza A viruses.”
Explore the full study: https://doi.org/10.1093/bioadv/vbaf254
Explore the full study: https://doi.org/10.1093/bioadv/vbaf254
October 23, 2025 at 10:01 AM
🦠 Now published in Bioinformatics Advances: “K-FluDB: A novel k-mer based database for enhanced genomic surveillance of influenza A viruses.”
Explore the full study: https://doi.org/10.1093/bioadv/vbaf254
Explore the full study: https://doi.org/10.1093/bioadv/vbaf254
🧩 Explore the latest from Bioinformatics Advances: “Nextpie: A web-based reporting tool and database for reproducible Nextflow pipelines.”
Read the full paper here: https://doi.org/10.1093/bioadv/vbaf252
Authors include: @lonnberg-lab.bsky.social
Read the full paper here: https://doi.org/10.1093/bioadv/vbaf252
Authors include: @lonnberg-lab.bsky.social

October 23, 2025 at 9:01 AM
🧩 Explore the latest from Bioinformatics Advances: “Nextpie: A web-based reporting tool and database for reproducible Nextflow pipelines.”
Read the full paper here: https://doi.org/10.1093/bioadv/vbaf252
Authors include: @lonnberg-lab.bsky.social
Read the full paper here: https://doi.org/10.1093/bioadv/vbaf252
Authors include: @lonnberg-lab.bsky.social
🧬 Just out in Bioinformatics Advances: “OncotreeVIS: An interactive graphical user interface for visualizing mutation tree cohorts.”
Explore the full study: https://doi.org/10.1093/bioadv/vbaf247
Authors include: @cbg.ethz.ch
Explore the full study: https://doi.org/10.1093/bioadv/vbaf247
Authors include: @cbg.ethz.ch

October 22, 2025 at 10:02 AM
🧬 Just out in Bioinformatics Advances: “OncotreeVIS: An interactive graphical user interface for visualizing mutation tree cohorts.”
Explore the full study: https://doi.org/10.1093/bioadv/vbaf247
Authors include: @cbg.ethz.ch
Explore the full study: https://doi.org/10.1093/bioadv/vbaf247
Authors include: @cbg.ethz.ch
🌍 Now published in Bioinformatics Advances: “GeoGenIE: A deep learning approach to predict geographic provenance of biodiversity samples from genomic SNPs.” Read the full paper here: https://doi.org/10.1093/bioadv/vbaf250

October 22, 2025 at 9:02 AM
🌍 Now published in Bioinformatics Advances: “GeoGenIE: A deep learning approach to predict geographic provenance of biodiversity samples from genomic SNPs.” Read the full paper here: https://doi.org/10.1093/bioadv/vbaf250
🧠 Explore the latest from Bioinformatics Advances: “KinMethyl: Robust methylation detection in prokaryotic SMRT sequencing via kinetic signal modeling and deep feature integration.”
Full article available: https://doi.org/10.1093/bioadv/vbaf249
Full article available: https://doi.org/10.1093/bioadv/vbaf249

October 21, 2025 at 10:01 AM
🧠 Explore the latest from Bioinformatics Advances: “KinMethyl: Robust methylation detection in prokaryotic SMRT sequencing via kinetic signal modeling and deep feature integration.”
Full article available: https://doi.org/10.1093/bioadv/vbaf249
Full article available: https://doi.org/10.1093/bioadv/vbaf249
🧪 Now published in Bioinformatics Advances: “Retention order prediction of peptides containing non-proteinogenic amino acids.”
Explore the full study: https://doi.org/10.1093/bioadv/vbaf246
Explore the full study: https://doi.org/10.1093/bioadv/vbaf246

October 21, 2025 at 9:02 AM
🧪 Now published in Bioinformatics Advances: “Retention order prediction of peptides containing non-proteinogenic amino acids.”
Explore the full study: https://doi.org/10.1093/bioadv/vbaf246
Explore the full study: https://doi.org/10.1093/bioadv/vbaf246

