
BioExcel CoE
@bioexcelcoe.bsky.social
Centre of Excellence for Computational Biomolecular Research
Save the dates!
The 3rd BioExcel Conference on Advances in Biomolecular Simulations is happening:
🗓️ 27-30 September 2026
🌐 Brno, Czech Republic
ℹ️ bioexcel.eu/hzr0
Registration opens: 19 January 2026
#moleculardynamics #freeenergy #drugdesign #ai #conference
The 3rd BioExcel Conference on Advances in Biomolecular Simulations is happening:
🗓️ 27-30 September 2026
🌐 Brno, Czech Republic
ℹ️ bioexcel.eu/hzr0
Registration opens: 19 January 2026
#moleculardynamics #freeenergy #drugdesign #ai #conference

November 10, 2025 at 3:21 PM
Save the dates!
The 3rd BioExcel Conference on Advances in Biomolecular Simulations is happening:
🗓️ 27-30 September 2026
🌐 Brno, Czech Republic
ℹ️ bioexcel.eu/hzr0
Registration opens: 19 January 2026
#moleculardynamics #freeenergy #drugdesign #ai #conference
The 3rd BioExcel Conference on Advances in Biomolecular Simulations is happening:
🗓️ 27-30 September 2026
🌐 Brno, Czech Republic
ℹ️ bioexcel.eu/hzr0
Registration opens: 19 January 2026
#moleculardynamics #freeenergy #drugdesign #ai #conference
Last chance to provide your feedback!
Our BioExcel 2025 survey is closing tomorrow, 31st October 🎃
Please fill in the survey ➡️bit.ly/3Vi7HE1
Our BioExcel 2025 survey is closing tomorrow, 31st October 🎃
Please fill in the survey ➡️bit.ly/3Vi7HE1

October 30, 2025 at 9:30 AM
Last chance to provide your feedback!
Our BioExcel 2025 survey is closing tomorrow, 31st October 🎃
Please fill in the survey ➡️bit.ly/3Vi7HE1
Our BioExcel 2025 survey is closing tomorrow, 31st October 🎃
Please fill in the survey ➡️bit.ly/3Vi7HE1
Our BioExcel 2025 survey is closing at the end of October ➡️bit.ly/3Vi7HE1
Please take this opportunity to provide us with your feedback!
Please take this opportunity to provide us with your feedback!

October 16, 2025 at 1:32 PM
Our BioExcel 2025 survey is closing at the end of October ➡️bit.ly/3Vi7HE1
Please take this opportunity to provide us with your feedback!
Please take this opportunity to provide us with your feedback!
Last chance to register ❗️
Our special edition webinar featuring talks on #topoisomerase inhibition, transmembrane #signalling and #membrane multilamellarity is this afternoon at 15:00 CEST
Register here ➡️bioexcel.eu/ion8
Our special edition webinar featuring talks on #topoisomerase inhibition, transmembrane #signalling and #membrane multilamellarity is this afternoon at 15:00 CEST
Register here ➡️bioexcel.eu/ion8

September 23, 2025 at 8:00 AM
Last chance to register ❗️
Our special edition webinar featuring talks on #topoisomerase inhibition, transmembrane #signalling and #membrane multilamellarity is this afternoon at 15:00 CEST
Register here ➡️bioexcel.eu/ion8
Our special edition webinar featuring talks on #topoisomerase inhibition, transmembrane #signalling and #membrane multilamellarity is this afternoon at 15:00 CEST
Register here ➡️bioexcel.eu/ion8
Happening next week!
Our 23-September, special edition webinar will feature talks on topoisomerase inhibition, transmembrane signalling and membrane multilamellarity
Register here ➡️bioexcel.eu/ion8
Our 23-September, special edition webinar will feature talks on topoisomerase inhibition, transmembrane signalling and membrane multilamellarity
Register here ➡️bioexcel.eu/ion8

September 17, 2025 at 11:53 AM
Happening next week!
Our 23-September, special edition webinar will feature talks on topoisomerase inhibition, transmembrane signalling and membrane multilamellarity
Register here ➡️bioexcel.eu/ion8
Our 23-September, special edition webinar will feature talks on topoisomerase inhibition, transmembrane signalling and membrane multilamellarity
Register here ➡️bioexcel.eu/ion8
5️⃣ Featuring the fifth of our showcase projects
Our automated pipeline, built on BioBBs, uses #AlphaFold, MD simulations, and virtual screening to output a list of ligand poses ➡️ bioexcel.eu/1c1j
#DrugDiscovery #Workflows #CADD #VirtualScreening
Our automated pipeline, built on BioBBs, uses #AlphaFold, MD simulations, and virtual screening to output a list of ligand poses ➡️ bioexcel.eu/1c1j
#DrugDiscovery #Workflows #CADD #VirtualScreening

September 12, 2025 at 3:17 PM
5️⃣ Featuring the fifth of our showcase projects
Our automated pipeline, built on BioBBs, uses #AlphaFold, MD simulations, and virtual screening to output a list of ligand poses ➡️ bioexcel.eu/1c1j
#DrugDiscovery #Workflows #CADD #VirtualScreening
Our automated pipeline, built on BioBBs, uses #AlphaFold, MD simulations, and virtual screening to output a list of ligand poses ➡️ bioexcel.eu/1c1j
#DrugDiscovery #Workflows #CADD #VirtualScreening
4️⃣ Featuring the fourth of our showcase projects
Upgrading GROMACS to handle billion-atom systems and enhancing I/O performance and precision, making the first-ever whole-cell simulation possible ➡️ bioexcel.eu/uw67
#MolecularDynamics #GROMACS #ComputationalBiology
Upgrading GROMACS to handle billion-atom systems and enhancing I/O performance and precision, making the first-ever whole-cell simulation possible ➡️ bioexcel.eu/uw67
#MolecularDynamics #GROMACS #ComputationalBiology

September 12, 2025 at 6:22 AM
4️⃣ Featuring the fourth of our showcase projects
Upgrading GROMACS to handle billion-atom systems and enhancing I/O performance and precision, making the first-ever whole-cell simulation possible ➡️ bioexcel.eu/uw67
#MolecularDynamics #GROMACS #ComputationalBiology
Upgrading GROMACS to handle billion-atom systems and enhancing I/O performance and precision, making the first-ever whole-cell simulation possible ➡️ bioexcel.eu/uw67
#MolecularDynamics #GROMACS #ComputationalBiology
3️⃣ Featuring the third of our showcase projects
We are tackling the tough-to-predict H3 antibody/nanobody loop using AlphaFlow and hierarchical clustering to create more accurate models ➡️bioexcel.eu/56jw
#Antibodies #AlphaFlow #DrugDiscovery
We are tackling the tough-to-predict H3 antibody/nanobody loop using AlphaFlow and hierarchical clustering to create more accurate models ➡️bioexcel.eu/56jw
#Antibodies #AlphaFlow #DrugDiscovery

September 11, 2025 at 1:18 PM
3️⃣ Featuring the third of our showcase projects
We are tackling the tough-to-predict H3 antibody/nanobody loop using AlphaFlow and hierarchical clustering to create more accurate models ➡️bioexcel.eu/56jw
#Antibodies #AlphaFlow #DrugDiscovery
We are tackling the tough-to-predict H3 antibody/nanobody loop using AlphaFlow and hierarchical clustering to create more accurate models ➡️bioexcel.eu/56jw
#Antibodies #AlphaFlow #DrugDiscovery
2⃣ Featuring the second of our showcase projects
Through a combination of AlphaFold2 and other tools we are bypassing the limits of traditional MD simulations ➡️ bioexcel.eu/dvy3
#Proteins #MolecularDynamics #GROMACS #AlphaFold2
Through a combination of AlphaFold2 and other tools we are bypassing the limits of traditional MD simulations ➡️ bioexcel.eu/dvy3
#Proteins #MolecularDynamics #GROMACS #AlphaFold2

September 9, 2025 at 8:04 PM
2⃣ Featuring the second of our showcase projects
Through a combination of AlphaFold2 and other tools we are bypassing the limits of traditional MD simulations ➡️ bioexcel.eu/dvy3
#Proteins #MolecularDynamics #GROMACS #AlphaFold2
Through a combination of AlphaFold2 and other tools we are bypassing the limits of traditional MD simulations ➡️ bioexcel.eu/dvy3
#Proteins #MolecularDynamics #GROMACS #AlphaFold2
1️⃣ Featuring the first of our showcase projects
We use an autoencoder to capture and quantify dynamic differences between protein conformational states ➡️ bioexcel.eu/wv4a
#MolecularDynamics #MachineLearning
We use an autoencoder to capture and quantify dynamic differences between protein conformational states ➡️ bioexcel.eu/wv4a
#MolecularDynamics #MachineLearning

September 8, 2025 at 9:03 PM
1️⃣ Featuring the first of our showcase projects
We use an autoencoder to capture and quantify dynamic differences between protein conformational states ➡️ bioexcel.eu/wv4a
#MolecularDynamics #MachineLearning
We use an autoencoder to capture and quantify dynamic differences between protein conformational states ➡️ bioexcel.eu/wv4a
#MolecularDynamics #MachineLearning
From support and training to which features we should develop in our core software, your views are valuable to us 💎
Our annual survey gives you the opportunity to provide us with all-round feedback
Please fill in the BioExcel 2025 survey ➡️bit.ly/3Vi7HE1
Our annual survey gives you the opportunity to provide us with all-round feedback
Please fill in the BioExcel 2025 survey ➡️bit.ly/3Vi7HE1

September 3, 2025 at 11:42 AM
From support and training to which features we should develop in our core software, your views are valuable to us 💎
Our annual survey gives you the opportunity to provide us with all-round feedback
Please fill in the BioExcel 2025 survey ➡️bit.ly/3Vi7HE1
Our annual survey gives you the opportunity to provide us with all-round feedback
Please fill in the BioExcel 2025 survey ➡️bit.ly/3Vi7HE1
📢Our next webinar is a special edition featuring the BioExcel Summer School 2025 poster prize winners
🗓️ Join us on 23 September at 15:00 CET
✍️ Registration and further information ➡️bioexcel.eu/ion8
Find out more about our speakers and their research...
🗓️ Join us on 23 September at 15:00 CET
✍️ Registration and further information ➡️bioexcel.eu/ion8
Find out more about our speakers and their research...

September 2, 2025 at 9:26 AM
📢Our next webinar is a special edition featuring the BioExcel Summer School 2025 poster prize winners
🗓️ Join us on 23 September at 15:00 CET
✍️ Registration and further information ➡️bioexcel.eu/ion8
Find out more about our speakers and their research...
🗓️ Join us on 23 September at 15:00 CET
✍️ Registration and further information ➡️bioexcel.eu/ion8
Find out more about our speakers and their research...
🤝 Our collaboration with other leading EU #HPC Centres of Excellence has resulted a white paper providing guidance and best practices for CoEs aiming to build their #innovation and #commercialisation capabilities
Read more: bioexcel.eu/k7b9
DOI: dx.doi.org/10.13140/RG....
Read more: bioexcel.eu/k7b9
DOI: dx.doi.org/10.13140/RG....

August 6, 2025 at 3:26 PM
🤝 Our collaboration with other leading EU #HPC Centres of Excellence has resulted a white paper providing guidance and best practices for CoEs aiming to build their #innovation and #commercialisation capabilities
Read more: bioexcel.eu/k7b9
DOI: dx.doi.org/10.13140/RG....
Read more: bioexcel.eu/k7b9
DOI: dx.doi.org/10.13140/RG....
👏 Congratulations to the Summer School 2025 poster prize winners!
🏆 Zuzana Janáčková
🏆 Jan van Elteren
🏆 Gesa Laura Freimann
Join us for a special edition webinar where they will present their work
🗓️ 23 September 2025, 15:00 CET
🏆 Zuzana Janáčková
🏆 Jan van Elteren
🏆 Gesa Laura Freimann
Join us for a special edition webinar where they will present their work
🗓️ 23 September 2025, 15:00 CET

June 16, 2025 at 1:21 PM
👏 Congratulations to the Summer School 2025 poster prize winners!
🏆 Zuzana Janáčková
🏆 Jan van Elteren
🏆 Gesa Laura Freimann
Join us for a special edition webinar where they will present their work
🗓️ 23 September 2025, 15:00 CET
🏆 Zuzana Janáčková
🏆 Jan van Elteren
🏆 Gesa Laura Freimann
Join us for a special edition webinar where they will present their work
🗓️ 23 September 2025, 15:00 CET
Today was the last day of our Summer School on Biomolecular Simulations 2025 ☀ 🏖
Finishing off with an introduction to QM/MM by Emiliano Ippoliti and a 'careers session' for all participants
Thank you to everyone for a great event 👋
Finishing off with an introduction to QM/MM by Emiliano Ippoliti and a 'careers session' for all participants
Thank you to everyone for a great event 👋

June 13, 2025 at 5:01 PM
Today was the last day of our Summer School on Biomolecular Simulations 2025 ☀ 🏖
Finishing off with an introduction to QM/MM by Emiliano Ippoliti and a 'careers session' for all participants
Thank you to everyone for a great event 👋
Finishing off with an introduction to QM/MM by Emiliano Ippoliti and a 'careers session' for all participants
Thank you to everyone for a great event 👋
During the afternoon session we were joined online by Sergey Ovchinnikov who gave some invaluable insights into #alphafold
Sergey talked about AlphaFold3 and its limitations. If you are looking for alternatives, check out ColabFold ➡ bit.ly/3KQEHOe
@sokrypton.org
Sergey talked about AlphaFold3 and its limitations. If you are looking for alternatives, check out ColabFold ➡ bit.ly/3KQEHOe
@sokrypton.org

June 12, 2025 at 3:40 PM
During the afternoon session we were joined online by Sergey Ovchinnikov who gave some invaluable insights into #alphafold
Sergey talked about AlphaFold3 and its limitations. If you are looking for alternatives, check out ColabFold ➡ bit.ly/3KQEHOe
@sokrypton.org
Sergey talked about AlphaFold3 and its limitations. If you are looking for alternatives, check out ColabFold ➡ bit.ly/3KQEHOe
@sokrypton.org
The tutorial uses #GROMACS as the simulation engine. Find this and other #workflow examples here ➡ bit.ly/3xzZJ0q
#BioBB enables automated system preparation, simulation and analysis, making your workflows and environment reproducible and #FAIR
#BioBB enables automated system preparation, simulation and analysis, making your workflows and environment reproducible and #FAIR

June 12, 2025 at 3:30 PM
The tutorial uses #GROMACS as the simulation engine. Find this and other #workflow examples here ➡ bit.ly/3xzZJ0q
#BioBB enables automated system preparation, simulation and analysis, making your workflows and environment reproducible and #FAIR
#BioBB enables automated system preparation, simulation and analysis, making your workflows and environment reproducible and #FAIR
Day 4 of the Summer School 2025 was about workflows and protein structure prediction
Adam Hospital introduced BioExcel Building Blocks #BioBB and followed up with a hands-on #tutorial on how to perform protein-ligand complex #moleculardynamics in Jupyter notebooks using BioBB
Adam Hospital introduced BioExcel Building Blocks #BioBB and followed up with a hands-on #tutorial on how to perform protein-ligand complex #moleculardynamics in Jupyter notebooks using BioBB

June 12, 2025 at 3:29 PM
Day 4 of the Summer School 2025 was about workflows and protein structure prediction
Adam Hospital introduced BioExcel Building Blocks #BioBB and followed up with a hands-on #tutorial on how to perform protein-ligand complex #moleculardynamics in Jupyter notebooks using BioBB
Adam Hospital introduced BioExcel Building Blocks #BioBB and followed up with a hands-on #tutorial on how to perform protein-ligand complex #moleculardynamics in Jupyter notebooks using BioBB
Sudarshan Behera gave an intro to free energy calculations followed by a #tutorial on using #pmx and #GROMACS to set up a ligand-binding relative free energy calculation study
Access the tutorial ➡ bit.ly/45Bcp3V and a #webinar on the topic ➡ bit.ly/3XjSJxf 📹
@sudars.bsky.social
Access the tutorial ➡ bit.ly/45Bcp3V and a #webinar on the topic ➡ bit.ly/3XjSJxf 📹
@sudars.bsky.social
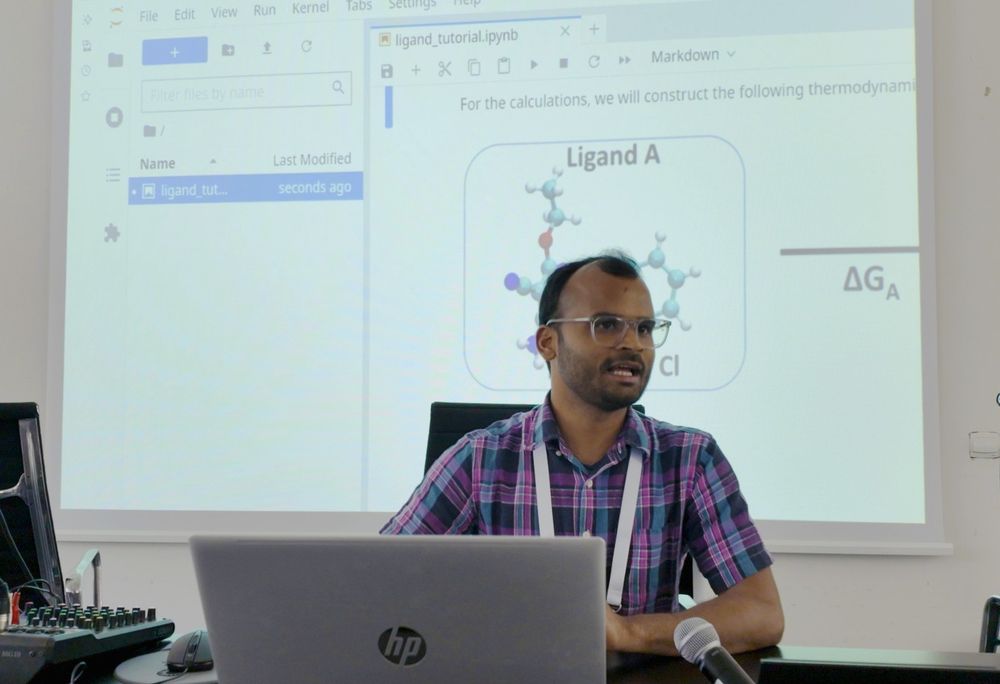
June 11, 2025 at 3:46 PM
Sudarshan Behera gave an intro to free energy calculations followed by a #tutorial on using #pmx and #GROMACS to set up a ligand-binding relative free energy calculation study
Access the tutorial ➡ bit.ly/45Bcp3V and a #webinar on the topic ➡ bit.ly/3XjSJxf 📹
@sudars.bsky.social
Access the tutorial ➡ bit.ly/45Bcp3V and a #webinar on the topic ➡ bit.ly/3XjSJxf 📹
@sudars.bsky.social
This was illustrated during the hands-on session on calculating the potential of mean force (PMF) along a reaction coordinate using the AWH method in #GROMACS
Find the #tutorial here ➡ bit.ly/4c8xeWG and our #webinar on its application ➡ bit.ly/3xqERbY 📽️
Find the #tutorial here ➡ bit.ly/4c8xeWG and our #webinar on its application ➡ bit.ly/3xqERbY 📽️

June 11, 2025 at 12:23 PM
This was illustrated during the hands-on session on calculating the potential of mean force (PMF) along a reaction coordinate using the AWH method in #GROMACS
Find the #tutorial here ➡ bit.ly/4c8xeWG and our #webinar on its application ➡ bit.ly/3xqERbY 📽️
Find the #tutorial here ➡ bit.ly/4c8xeWG and our #webinar on its application ➡ bit.ly/3xqERbY 📽️
Day 3 of the Summer School is about enhanced sampling and free energy calculations⚡️
Berk Hess introduced the AWH method implemented in #GROMACS and illustrated its application to alchemical transformations where the main challenge is to define a good reaction coordinate
Berk Hess introduced the AWH method implemented in #GROMACS and illustrated its application to alchemical transformations where the main challenge is to define a good reaction coordinate

June 11, 2025 at 12:22 PM
Day 3 of the Summer School is about enhanced sampling and free energy calculations⚡️
Berk Hess introduced the AWH method implemented in #GROMACS and illustrated its application to alchemical transformations where the main challenge is to define a good reaction coordinate
Berk Hess introduced the AWH method implemented in #GROMACS and illustrated its application to alchemical transformations where the main challenge is to define a good reaction coordinate
An engaging tutorial by Attilio, Giuliano Malloci and Mohd Athar enabled participants to test the #EDESmethod for enhanced sampling ➡ bit.ly/45Rdp3h

June 10, 2025 at 5:05 PM
An engaging tutorial by Attilio, Giuliano Malloci and Mohd Athar enabled participants to test the #EDESmethod for enhanced sampling ➡ bit.ly/45Rdp3h
Attilio Vargiu gave an in-depth lecture on biomolecular recognition and enhanced sampling methods, covering techniques like Replica Exchange MD, Metadynamics, Accelerated MD, Targeted MD, and more, illustrated with multiple case studies

June 10, 2025 at 5:05 PM
Attilio Vargiu gave an in-depth lecture on biomolecular recognition and enhanced sampling methods, covering techniques like Replica Exchange MD, Metadynamics, Accelerated MD, Targeted MD, and more, illustrated with multiple case studies
Anna Kravchenko took participants through the HADDOCK3 tutorial for antibody-antigen modeling
This tutorial and other useful learnings including how to carry out antibody-antigen structure prediction from sequence using #alphafold can be found here ➡ bit.ly/45LCWgY
This tutorial and other useful learnings including how to carry out antibody-antigen structure prediction from sequence using #alphafold can be found here ➡ bit.ly/45LCWgY

June 10, 2025 at 1:43 PM
Anna Kravchenko took participants through the HADDOCK3 tutorial for antibody-antigen modeling
This tutorial and other useful learnings including how to carry out antibody-antigen structure prediction from sequence using #alphafold can be found here ➡ bit.ly/45LCWgY
This tutorial and other useful learnings including how to carry out antibody-antigen structure prediction from sequence using #alphafold can be found here ➡ bit.ly/45LCWgY
Day 2 of the Summer School is about molecular recognition and docking 🤝
@amjjbonvin.bsky.social introduces #HADDOCK to predict interactions between: proteins, nucleic acids, glycans and small molecules
For further flexibility and customisation check out HADDOCK3 ➡️bit.ly/4mQLZn7
@amjjbonvin.bsky.social introduces #HADDOCK to predict interactions between: proteins, nucleic acids, glycans and small molecules
For further flexibility and customisation check out HADDOCK3 ➡️bit.ly/4mQLZn7

June 10, 2025 at 12:56 PM
Day 2 of the Summer School is about molecular recognition and docking 🤝
@amjjbonvin.bsky.social introduces #HADDOCK to predict interactions between: proteins, nucleic acids, glycans and small molecules
For further flexibility and customisation check out HADDOCK3 ➡️bit.ly/4mQLZn7
@amjjbonvin.bsky.social introduces #HADDOCK to predict interactions between: proteins, nucleic acids, glycans and small molecules
For further flexibility and customisation check out HADDOCK3 ➡️bit.ly/4mQLZn7

