At the Repetitive DNA Biology (REPBIO) Lab, we leverage the latest technologies to decode nucleotide sequences for investigating how repetitive DNA shapes genome function and contributes to disease.






Microhomology patterns observed at 5´ inversion and truncation breakpoint junctions for 1.271 L1 insertions point to MMEJ. We also found three templated insertions, suggestive of polymerase theta. [3/9]
www.cell.com/cell/fulltex...

Microhomology patterns observed at 5´ inversion and truncation breakpoint junctions for 1.271 L1 insertions point to MMEJ. We also found three templated insertions, suggestive of polymerase theta. [3/9]
www.cell.com/cell/fulltex...
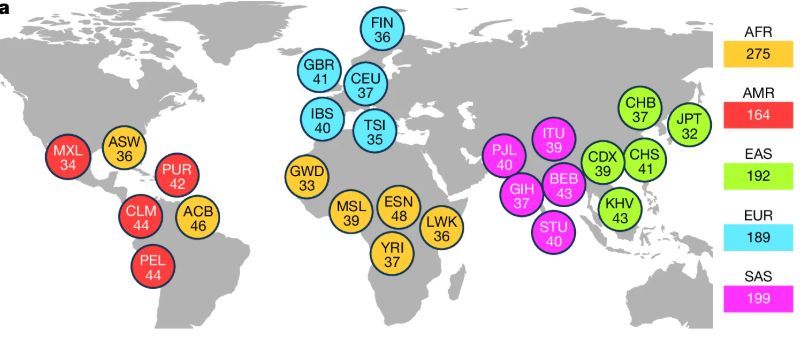
MEI analysis on 32 haplotype-resolved human genomes. Older L1s have higher allele frequencies, lower ORF preservation and are therefore less active, but eternal youth can occur [2/9]
www.science.org/doi/10.1126/...

MEI analysis on 32 haplotype-resolved human genomes. Older L1s have higher allele frequencies, lower ORF preservation and are therefore less active, but eternal youth can occur [2/9]
www.science.org/doi/10.1126/...