
Group Leader at Barts Cancer Institute in London. Working on somatic evolution, ecDNA, clonal haematopoiesis, gITH, clonal evolution…
https://www.bartscancer.london/staff/dr-benjamin-werner/
Contact: b.werner@qmul.ac.uk

Computational modelling suggests the following scenario:
First ecDNA forms and accumulates in pre-cancerous cells. Later one ecDNA copy acquires the variant and possibly starts the clonal expansion
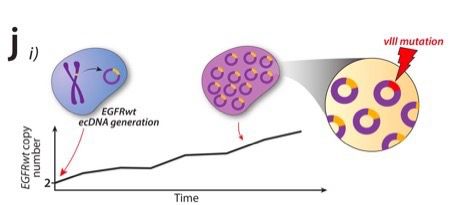
Computational modelling suggests the following scenario:
First ecDNA forms and accumulates in pre-cancerous cells. Later one ecDNA copy acquires the variant and possibly starts the clonal expansion

But interestingly we found a few EGFR variants exclusively amplified on ecDNA (EGFRvIII and others) that seem under strong positive selection but were always ecDNA subclonal

But interestingly we found a few EGFR variants exclusively amplified on ecDNA (EGFRvIII and others) that seem under strong positive selection but were always ecDNA subclonal






To make sure you never learn conversion of cm and inches 😂

To make sure you never learn conversion of cm and inches 😂

Many thanks to @cancerevo.org and @zaccasimo.bsky.social to be examiners!

Many thanks to @cancerevo.org and @zaccasimo.bsky.social to be examiners!











