Axel Visel
@axelvisel.bsky.social
Genome Scientist. Studies how DNA makes humans, mice, plants, microbes. At Lawrence Berkeley National Lab and Joint Genome Institute. Views are my own.
To find the right email address, go to PubMed and expand the "Affiliations". It's usually the last author (for biology articles).
September 23, 2025 at 1:23 PM
To find the right email address, go to PubMed and expand the "Affiliations". It's usually the last author (for biology articles).
Looks like I made the puzzle too difficult. It's been a month and still no reports of anyone having found all 10.
Which confirms: Enhancer discovery is hard!
Which confirms: Enhancer discovery is hard!
September 12, 2025 at 3:37 AM
Looks like I made the puzzle too difficult. It's been a month and still no reports of anyone having found all 10.
Which confirms: Enhancer discovery is hard!
Which confirms: Enhancer discovery is hard!
Here is our speaker lineup:
jgi.doe.gov/work-with-us...
Featuring @nekton4plankton.bsky.social @annedekas.bsky.social @microyunha.bsky.social @masarunobu.bsky.social and more
jgi.doe.gov/work-with-us...
Featuring @nekton4plankton.bsky.social @annedekas.bsky.social @microyunha.bsky.social @masarunobu.bsky.social and more

August 25, 2025 at 9:47 PM
Here is our speaker lineup:
jgi.doe.gov/work-with-us...
Featuring @nekton4plankton.bsky.social @annedekas.bsky.social @microyunha.bsky.social @masarunobu.bsky.social and more
jgi.doe.gov/work-with-us...
Featuring @nekton4plankton.bsky.social @annedekas.bsky.social @microyunha.bsky.social @masarunobu.bsky.social and more
If you are interested in using DAPseq for your plant, algal, fungal or microbial genomes, consider applying to one of @jgi.doe.gov's user programs:
jgi.doe.gov/work-with-us...
jgi.doe.gov/work-with-us...

User Programs | Joint Genome Institute
Learn more about our Community Science Program, as well as other collaborative opportunities available through the FICUS call and other special initiatives.
jgi.doe.gov
August 20, 2025 at 12:53 AM
If you are interested in using DAPseq for your plant, algal, fungal or microbial genomes, consider applying to one of @jgi.doe.gov's user programs:
jgi.doe.gov/work-with-us...
jgi.doe.gov/work-with-us...
Also see this related News and Views by Mary Galli and Andrea Gallavotti
www.nature.com/articles/s41...
www.nature.com/articles/s41...

A binding agreement - Nature Plants
Transcription factors are proteins that recognize and bind short, specific DNA sequences and regulate when, where and how genes are expressed in a cell. A recent large-scale study of transcription fac...
www.nature.com
August 20, 2025 at 12:48 AM
Also see this related News and Views by Mary Galli and Andrea Gallavotti
www.nature.com/articles/s41...
www.nature.com/articles/s41...
Yes, absolutely! They are clearly not all artifacts. But some of them are and the problem is more pronounced in plants than in animals. Fortunately, the preprint also highlights how a unified re-annotation strategy can reduce these artifacts, making whatever PAV remains even more interesting!

August 18, 2025 at 7:31 PM
Yes, absolutely! They are clearly not all artifacts. But some of them are and the problem is more pronounced in plants than in animals. Fortunately, the preprint also highlights how a unified re-annotation strategy can reduce these artifacts, making whatever PAV remains even more interesting!
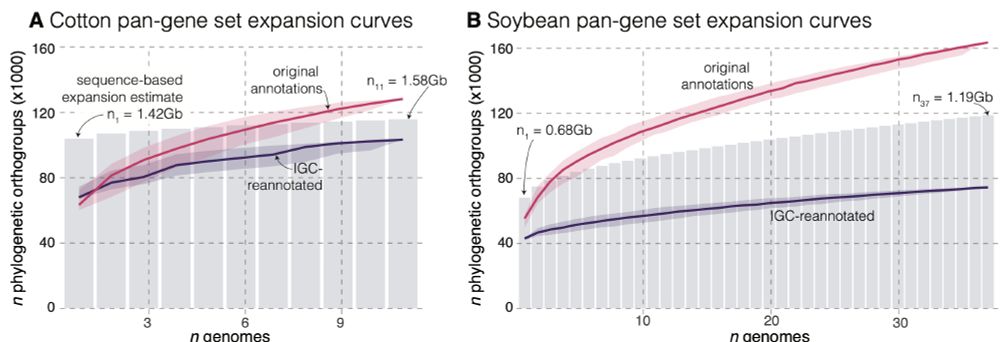
Determining presence-absence variation (PAV) across reference genomes is a major goal of pangenome analysis. It turns out that A LOT of gene PAV is due to methodological artifacts.
We explore the causes of this in soybean and cotton datasets in our recent preprint: www.biorxiv.org/content/10.1...
We explore the causes of this in soybean and cotton datasets in our recent preprint: www.biorxiv.org/content/10.1...
August 18, 2025 at 7:08 PM


