Axel Visel
@axelvisel.bsky.social
Genome Scientist. Studies how DNA makes humans, mice, plants, microbes. At Lawrence Berkeley National Lab and Joint Genome Institute. Views are my own.
Here is our speaker lineup:
jgi.doe.gov/work-with-us...
Featuring @nekton4plankton.bsky.social @annedekas.bsky.social @microyunha.bsky.social @masarunobu.bsky.social and more
jgi.doe.gov/work-with-us...
Featuring @nekton4plankton.bsky.social @annedekas.bsky.social @microyunha.bsky.social @masarunobu.bsky.social and more

August 25, 2025 at 9:47 PM
Here is our speaker lineup:
jgi.doe.gov/work-with-us...
Featuring @nekton4plankton.bsky.social @annedekas.bsky.social @microyunha.bsky.social @masarunobu.bsky.social and more
jgi.doe.gov/work-with-us...
Featuring @nekton4plankton.bsky.social @annedekas.bsky.social @microyunha.bsky.social @masarunobu.bsky.social and more
Postdoc opportunity with @tomasbruna.bsky.social here at @jgi.doe.gov @berkeleylab.lbl.gov:
Develop, benchmark, and apply Plant Genomic Language Models (gLMs) 🌿🧬💻
jobs.lbl.gov/jobs/computa...
#AI #PlantGenomics #gLMs #Postdoc
Develop, benchmark, and apply Plant Genomic Language Models (gLMs) 🌿🧬💻
jobs.lbl.gov/jobs/computa...
#AI #PlantGenomics #gLMs #Postdoc

August 21, 2025 at 12:05 AM
Postdoc opportunity with @tomasbruna.bsky.social here at @jgi.doe.gov @berkeleylab.lbl.gov:
Develop, benchmark, and apply Plant Genomic Language Models (gLMs) 🌿🧬💻
jobs.lbl.gov/jobs/computa...
#AI #PlantGenomics #gLMs #Postdoc
Develop, benchmark, and apply Plant Genomic Language Models (gLMs) 🌿🧬💻
jobs.lbl.gov/jobs/computa...
#AI #PlantGenomics #gLMs #Postdoc
On the cover of @natplants.nature.com this month:
@leobaumgart.bsky.social @greensi.bsky.social @abmora.bsky.social @omalley-regulome.bsky.social et al. map binding sites for 360 TFs across 10 🌿plant🌿 species using a new multiDAP approach
Here is Sharon's explainer 🧵:
bsky.app/profile/gree...
@leobaumgart.bsky.social @greensi.bsky.social @abmora.bsky.social @omalley-regulome.bsky.social et al. map binding sites for 360 TFs across 10 🌿plant🌿 species using a new multiDAP approach
Here is Sharon's explainer 🧵:
bsky.app/profile/gree...

August 20, 2025 at 12:45 AM
On the cover of @natplants.nature.com this month:
@leobaumgart.bsky.social @greensi.bsky.social @abmora.bsky.social @omalley-regulome.bsky.social et al. map binding sites for 360 TFs across 10 🌿plant🌿 species using a new multiDAP approach
Here is Sharon's explainer 🧵:
bsky.app/profile/gree...
@leobaumgart.bsky.social @greensi.bsky.social @abmora.bsky.social @omalley-regulome.bsky.social et al. map binding sites for 360 TFs across 10 🌿plant🌿 species using a new multiDAP approach
Here is Sharon's explainer 🧵:
bsky.app/profile/gree...
In 2026, @jgi.doe.gov and EMSL will (for the first time!) hold a joint user meeting in Seattle from March 3-5
Program details to be announced, but our registration site just launched - so you can already secure your spot:
jgi.doe.gov/work-with-us...
Program details to be announced, but our registration site just launched - so you can already secure your spot:
jgi.doe.gov/work-with-us...

August 19, 2025 at 5:30 PM
In 2026, @jgi.doe.gov and EMSL will (for the first time!) hold a joint user meeting in Seattle from March 3-5
Program details to be announced, but our registration site just launched - so you can already secure your spot:
jgi.doe.gov/work-with-us...
Program details to be announced, but our registration site just launched - so you can already secure your spot:
jgi.doe.gov/work-with-us...
Yes, absolutely! They are clearly not all artifacts. But some of them are and the problem is more pronounced in plants than in animals. Fortunately, the preprint also highlights how a unified re-annotation strategy can reduce these artifacts, making whatever PAV remains even more interesting!

August 18, 2025 at 7:31 PM
Yes, absolutely! They are clearly not all artifacts. But some of them are and the problem is more pronounced in plants than in animals. Fortunately, the preprint also highlights how a unified re-annotation strategy can reduce these artifacts, making whatever PAV remains even more interesting!
🌿Plant pangenomes🌿 have a LOT of genomic presence-absence-variation.
Or do they???
A new @jgi.doe.gov preprint by @tomasbruna.bsky.social @jotlovell.bsky.social @avril-m-harder.bsky.social Avinash Sreedasyam takes a closer look
www.biorxiv.org/content/10.1...
Or do they???
A new @jgi.doe.gov preprint by @tomasbruna.bsky.social @jotlovell.bsky.social @avril-m-harder.bsky.social Avinash Sreedasyam takes a closer look
www.biorxiv.org/content/10.1...

August 18, 2025 at 6:28 PM
🌿Plant pangenomes🌿 have a LOT of genomic presence-absence-variation.
Or do they???
A new @jgi.doe.gov preprint by @tomasbruna.bsky.social @jotlovell.bsky.social @avril-m-harder.bsky.social Avinash Sreedasyam takes a closer look
www.biorxiv.org/content/10.1...
Or do they???
A new @jgi.doe.gov preprint by @tomasbruna.bsky.social @jotlovell.bsky.social @avril-m-harder.bsky.social Avinash Sreedasyam takes a closer look
www.biorxiv.org/content/10.1...
Congratulations to Uma Grover on an outstanding poster presentation! 🎉
Uma joined @b-coli.bsky.social's and my group at @jgi.doe.gov @biosci.lbl.gov this summer as a @berkeleylab.lbl.gov SULI intern and carried out exciting work on single-cell profiling of plant-fungal interactions.
Uma joined @b-coli.bsky.social's and my group at @jgi.doe.gov @biosci.lbl.gov this summer as a @berkeleylab.lbl.gov SULI intern and carried out exciting work on single-cell profiling of plant-fungal interactions.

August 9, 2025 at 1:25 PM
Congratulations to Uma Grover on an outstanding poster presentation! 🎉
Uma joined @b-coli.bsky.social's and my group at @jgi.doe.gov @biosci.lbl.gov this summer as a @berkeleylab.lbl.gov SULI intern and carried out exciting work on single-cell profiling of plant-fungal interactions.
Uma joined @b-coli.bsky.social's and my group at @jgi.doe.gov @biosci.lbl.gov this summer as a @berkeleylab.lbl.gov SULI intern and carried out exciting work on single-cell profiling of plant-fungal interactions.
August 8, 2025 at 6:25 PM
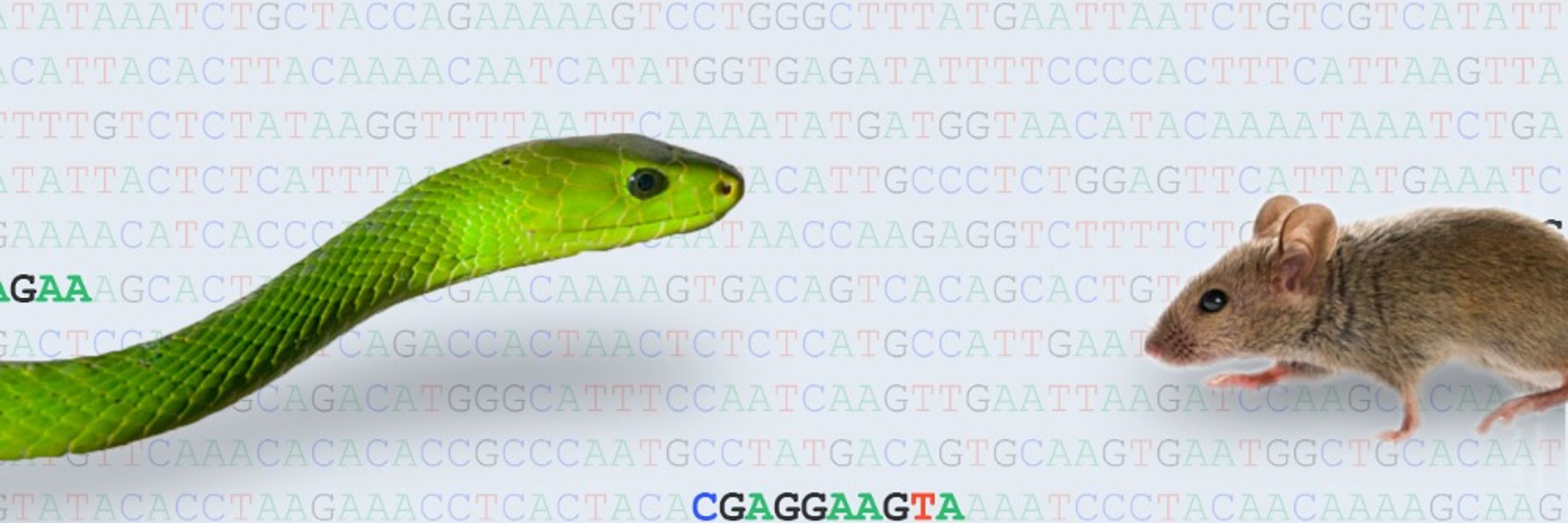
Hiding in plain sight - how close are we to mapping ALL 🧬enhancers🧬 in the genome?
Our new paper by Mannion et al. takes a systematic look at "hidden enhancers" and why they remain so hard to find. With @mosterwalder.bsky.social, @jlopezrios.bsky.social & many more
www.nature.com/articles/s41...
Our new paper by Mannion et al. takes a systematic look at "hidden enhancers" and why they remain so hard to find. With @mosterwalder.bsky.social, @jlopezrios.bsky.social & many more
www.nature.com/articles/s41...

August 8, 2025 at 6:10 PM
Hiding in plain sight - how close are we to mapping ALL 🧬enhancers🧬 in the genome?
Our new paper by Mannion et al. takes a systematic look at "hidden enhancers" and why they remain so hard to find. With @mosterwalder.bsky.social, @jlopezrios.bsky.social & many more
www.nature.com/articles/s41...
Our new paper by Mannion et al. takes a systematic look at "hidden enhancers" and why they remain so hard to find. With @mosterwalder.bsky.social, @jlopezrios.bsky.social & many more
www.nature.com/articles/s41...
🧬 @jgi.doe.gov is hiring! We're seeking an Advanced Analysis Manager to lead data infrastructure and software engineering supporting the generation, analysis, and dissemination of genomics data
Hybrid | SF Bay Area | Apply by Aug 7
jobs.lbl.gov/jobs/advance...
@biosci.lbl.gov @berkeleylab.lbl.gov
Hybrid | SF Bay Area | Apply by Aug 7
jobs.lbl.gov/jobs/advance...
@biosci.lbl.gov @berkeleylab.lbl.gov

July 29, 2025 at 11:49 PM
🧬 @jgi.doe.gov is hiring! We're seeking an Advanced Analysis Manager to lead data infrastructure and software engineering supporting the generation, analysis, and dissemination of genomics data
Hybrid | SF Bay Area | Apply by Aug 7
jobs.lbl.gov/jobs/advance...
@biosci.lbl.gov @berkeleylab.lbl.gov
Hybrid | SF Bay Area | Apply by Aug 7
jobs.lbl.gov/jobs/advance...
@biosci.lbl.gov @berkeleylab.lbl.gov
Catalan got it right, though:
"ratpenat" = the "feathered rat"
There is even one in Valencia's coat of arms
"ratpenat" = the "feathered rat"
There is even one in Valencia's coat of arms

July 16, 2025 at 6:48 PM
Catalan got it right, though:
"ratpenat" = the "feathered rat"
There is even one in Valencia's coat of arms
"ratpenat" = the "feathered rat"
There is even one in Valencia's coat of arms
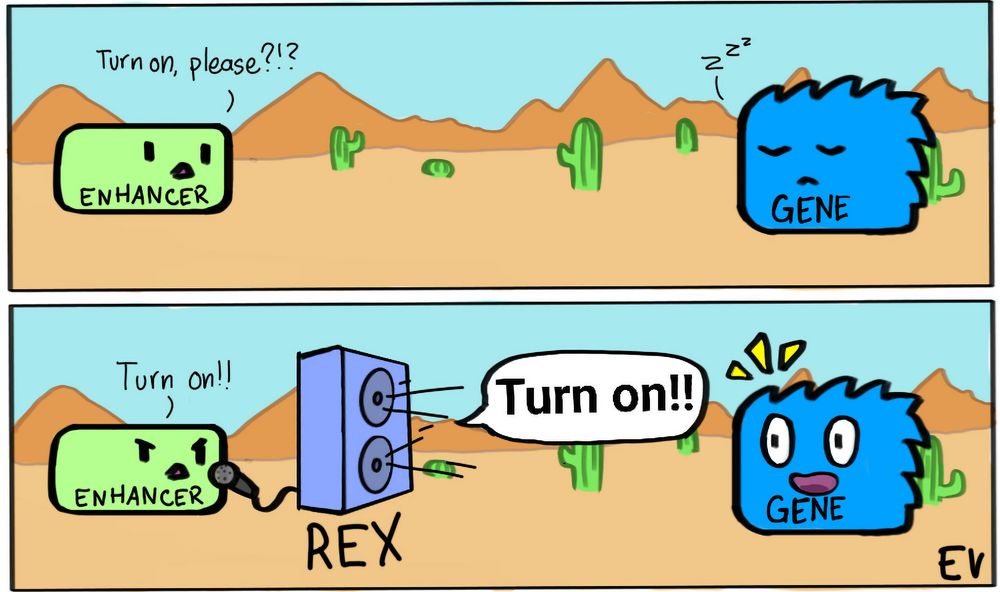
Getting the message across from a distance:
🧬REX🧬 is a “Range EXtender” element that can turn short-range into looooooooooooong-range enhancers
Great collaboration led by @gracebower.bsky.social and @evgenykvon.bsky.social
www.nature.com/articles/s41...
@biosci.lbl.gov @berkeleylab.lbl.gov
🧬REX🧬 is a “Range EXtender” element that can turn short-range into looooooooooooong-range enhancers
Great collaboration led by @gracebower.bsky.social and @evgenykvon.bsky.social
www.nature.com/articles/s41...
@biosci.lbl.gov @berkeleylab.lbl.gov
July 2, 2025 at 4:03 PM
Getting the message across from a distance:
🧬REX🧬 is a “Range EXtender” element that can turn short-range into looooooooooooong-range enhancers
Great collaboration led by @gracebower.bsky.social and @evgenykvon.bsky.social
www.nature.com/articles/s41...
@biosci.lbl.gov @berkeleylab.lbl.gov
🧬REX🧬 is a “Range EXtender” element that can turn short-range into looooooooooooong-range enhancers
Great collaboration led by @gracebower.bsky.social and @evgenykvon.bsky.social
www.nature.com/articles/s41...
@biosci.lbl.gov @berkeleylab.lbl.gov
💯Forgot to cover that in the thread. Yes, it's remarkable how well it worked with chromatin accessibility data alone (and to think how much better it might work with more data modalities and using single-cell data).

June 18, 2025 at 7:40 PM
💯Forgot to cover that in the thread. Yes, it's remarkable how well it worked with chromatin accessibility data alone (and to think how much better it might work with more data modalities and using single-cell data).
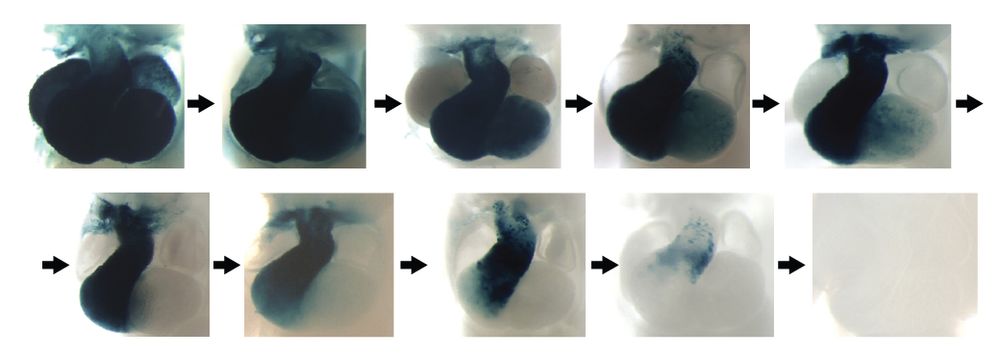
With respect to the “inner logic” of enhancers: Some enhancers were uniformly (across all structures) affected by loss-of-function mutations, while others had structure-specific motifs for subregions. Shown here for a heart enhancer.
June 18, 2025 at 6:09 PM
With respect to the “inner logic” of enhancers: Some enhancers were uniformly (across all structures) affected by loss-of-function mutations, while others had structure-specific motifs for subregions. Shown here for a heart enhancer.
Now the real question: Can the best-fit models correctly predict consequences of 1bp mutations? Yes - at least in several cases we tested. This has important implications for interpreting noncoding variants observed in clinical genetics data.

June 18, 2025 at 6:06 PM
Now the real question: Can the best-fit models correctly predict consequences of 1bp mutations? Yes - at least in several cases we tested. This has important implications for interpreting noncoding variants observed in clinical genetics data.
To further tweak this, we found that in some cases combining models from different tissues and/or considering degenerate (suboptimal) motifs improved prediction performance.

June 18, 2025 at 6:05 PM
To further tweak this, we found that in some cases combining models from different tissues and/or considering degenerate (suboptimal) motifs improved prediction performance.
We built models from relevant epigenomic data and used our in vivo data to benchmark their performance. We found that the DL models tell us the truth, but not the whole truth: Most predicted functional sites are real (PPV 88%), but they still miss ~40% of functional sites.

June 18, 2025 at 6:04 PM
We built models from relevant epigenomic data and used our in vivo data to benchmark their performance. We found that the DL models tell us the truth, but not the whole truth: Most predicted functional sites are real (PPV 88%), but they still miss ~40% of functional sites.
But who cares about blocks? We need generalizable, basepair-resolution predictions and insights about TF motifs. Enter @anshulkundaje.bsky.social @anusri.bsky.social and their ChromBPNet deep learning model of chromatin accessibility.
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

June 18, 2025 at 6:04 PM
But who cares about blocks? We need generalizable, basepair-resolution predictions and insights about TF motifs. Enter @anshulkundaje.bsky.social @anusri.bsky.social and their ChromBPNet deep learning model of chromatin accessibility.
www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...
But the overall sensitivity varied widely across the enhancers. One was hypersensitive - every block mattered. Most had “Achilles’ heel” blocks - single 12bp hits that wiped out function entirely. Only three had gain-of-function blocks, presumably corresponding to repressor sites.

June 18, 2025 at 6:02 PM
But the overall sensitivity varied widely across the enhancers. One was hypersensitive - every block mattered. Most had “Achilles’ heel” blocks - single 12bp hits that wiped out function entirely. Only three had gain-of-function blocks, presumably corresponding to repressor sites.
Turns out functional sequence features are scattered across the entire length of enhancers, and there are MANY of them. About two thirds of all blocks caused changes in function - mostly losses, but also some gains, i.e. ectopic enhancer activity in the wrong tissue.

June 18, 2025 at 6:02 PM
Turns out functional sequence features are scattered across the entire length of enhancers, and there are MANY of them. About two thirds of all blocks caused changes in function - mostly losses, but also some gains, i.e. ectopic enhancer activity in the wrong tissue.
Why 12bp blocks? The combined enhancers covered >2kb. That would require a LOT of experiments with 1bp mutations. Using block mutatagenesis, we covered all basepairs efficiently. It was still a LOT of experiments.

June 18, 2025 at 6:01 PM
Why 12bp blocks? The combined enhancers covered >2kb. That would require a LOT of experiments with 1bp mutations. Using block mutatagenesis, we covered all basepairs efficiently. It was still a LOT of experiments.
To find out, we systematically mutagenized 7 human enhancers in 12bp blocks and checked if these mutations change enhancer activity in developing mouse embryos.

June 18, 2025 at 6:00 PM
To find out, we systematically mutagenized 7 human enhancers in 12bp blocks and checked if these mutations change enhancer activity in developing mouse embryos.
Textbook knowledge: Enhancers consist of transcription factor binding sites (TFBSs). Once the right TFs have bound, the enhancer turns on.
But which of these sites are actually important? Do we know all the sites? What about redundant sites? Questions over questions.
But which of these sites are actually important? Do we know all the sites? What about redundant sites? Questions over questions.

June 18, 2025 at 5:58 PM
Textbook knowledge: Enhancers consist of transcription factor binding sites (TFBSs). Once the right TFs have bound, the enhancer turns on.
But which of these sites are actually important? Do we know all the sites? What about redundant sites? Questions over questions.
But which of these sites are actually important? Do we know all the sites? What about redundant sites? Questions over questions.
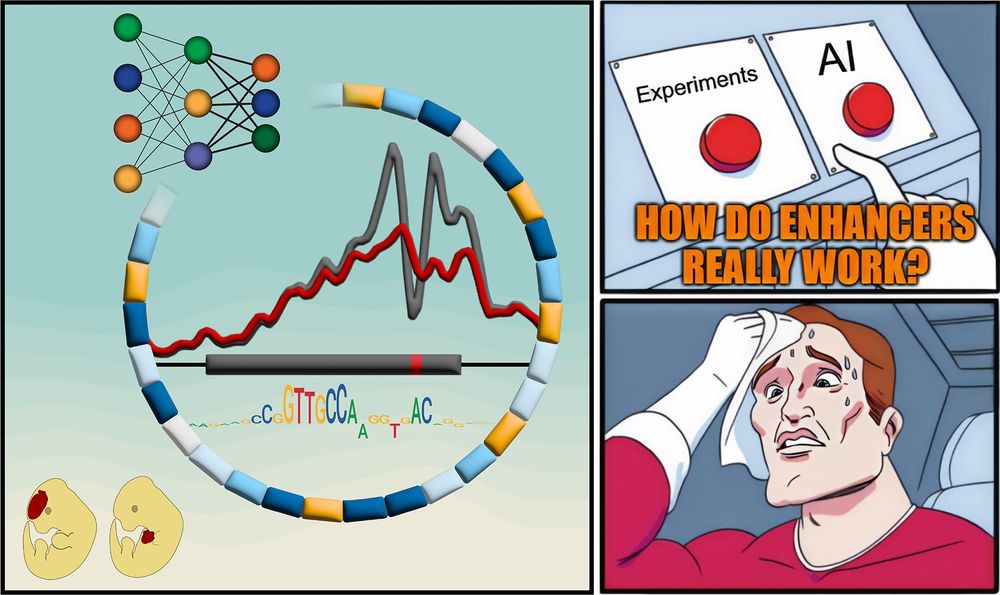
Textbooks: “Enhancers are just a bunch of TFBSs”
But how do they REALLY work?
New paper with many contributors here @berkeleylab.lbl.gov, @anshulkundaje.bsky.social, @anusri.bsky.social
A 🧵 (1/n)
Free access link: rdcu.be/erD22
But how do they REALLY work?
New paper with many contributors here @berkeleylab.lbl.gov, @anshulkundaje.bsky.social, @anusri.bsky.social
A 🧵 (1/n)
Free access link: rdcu.be/erD22
June 18, 2025 at 5:56 PM
Textbooks: “Enhancers are just a bunch of TFBSs”
But how do they REALLY work?
New paper with many contributors here @berkeleylab.lbl.gov, @anshulkundaje.bsky.social, @anusri.bsky.social
A 🧵 (1/n)
Free access link: rdcu.be/erD22
But how do they REALLY work?
New paper with many contributors here @berkeleylab.lbl.gov, @anshulkundaje.bsky.social, @anusri.bsky.social
A 🧵 (1/n)
Free access link: rdcu.be/erD22


