
Assaf Zaritsky
@assafzaritsky.bsky.social
Computational cell biologist
https://www.assafzaritsky.com/
https://www.assafzaritsky.com/
Somehow missed acknowledging Alon Shpigler, the first author 😱
October 30, 2025 at 2:15 PM
Somehow missed acknowledging Alon Shpigler, the first author 😱
Congratulations to Naor Kolet who led this project and thanks to co-authors Shahar Golan for his contribution and to @erinweisbart.bsky.social l for her critical insights regarding interpretability 🎉
3/3
3/3

October 30, 2025 at 9:44 AM
Congratulations to Naor Kolet who led this project and thanks to co-authors Shahar Golan for his contribution and to @erinweisbart.bsky.social l for her critical insights regarding interpretability 🎉
3/3
3/3
Anomaly-based representations have several benefits:
1. Reducing batch effects
2. Improving reproducibility and mechanism of action classification
3. Complementing classical representations
4. Revealing alterations in biologically interpretable dependencies
2/3
1. Reducing batch effects
2. Improving reproducibility and mechanism of action classification
3. Complementing classical representations
4. Revealing alterations in biologically interpretable dependencies
2/3
October 30, 2025 at 9:44 AM
Anomaly-based representations have several benefits:
1. Reducing batch effects
2. Improving reproducibility and mechanism of action classification
3. Complementing classical representations
4. Revealing alterations in biologically interpretable dependencies
2/3
1. Reducing batch effects
2. Improving reproducibility and mechanism of action classification
3. Complementing classical representations
4. Revealing alterations in biologically interpretable dependencies
2/3
Jennifer Lippincott-Schwartz, Emma Lundberg, Alex Mogilner, Maddy Parsons, @loicaroyer.bsky.social, Guillaume Salbreux, Anđela Šarić, Timm Schroeder, Hervé Turlier, @virginieuhlmann.bsky.social, Vincenzo Vitelli
Please save the date and spread the word!
3/3
Please save the date and spread the word!
3/3
October 10, 2025 at 9:23 AM
Jennifer Lippincott-Schwartz, Emma Lundberg, Alex Mogilner, Maddy Parsons, @loicaroyer.bsky.social, Guillaume Salbreux, Anđela Šarić, Timm Schroeder, Hervé Turlier, @virginieuhlmann.bsky.social, Vincenzo Vitelli
Please save the date and spread the word!
3/3
Please save the date and spread the word!
3/3
Co-organized with Meghan Driscoll, @quantumjot.bsky.social and Susanne Rafelski
Speakers include: @kevin-dean.bsky.social, @ellenberglab.bsky.social, Margaret Gardel, @florianjug.bsky.social, @krishnaswamylab.bsky.social, @levayerr.bsky.social, @priscaliberali.bsky.social, ...
Speakers include: @kevin-dean.bsky.social, @ellenberglab.bsky.social, Margaret Gardel, @florianjug.bsky.social, @krishnaswamylab.bsky.social, @levayerr.bsky.social, @priscaliberali.bsky.social, ...
October 10, 2025 at 9:23 AM
Co-organized with Meghan Driscoll, @quantumjot.bsky.social and Susanne Rafelski
Speakers include: @kevin-dean.bsky.social, @ellenberglab.bsky.social, Margaret Gardel, @florianjug.bsky.social, @krishnaswamylab.bsky.social, @levayerr.bsky.social, @priscaliberali.bsky.social, ...
Speakers include: @kevin-dean.bsky.social, @ellenberglab.bsky.social, Margaret Gardel, @florianjug.bsky.social, @krishnaswamylab.bsky.social, @levayerr.bsky.social, @priscaliberali.bsky.social, ...
Sorry, I meant @leeat-keren.bsky.social and Yael Amitay....
May 9, 2025 at 9:15 AM
Sorry, I meant @leeat-keren.bsky.social and Yael Amitay....
A creative and technically challenging idea led by @zamir_amos, with key contributions from Yuval Tamir and @yaelam75. This would not have been possible without the great collaboration with @leeat_keren! And thanks to @WellcomeLeap ΔTissue for funding!
16/n
16/n

May 9, 2025 at 9:08 AM
A creative and technically challenging idea led by @zamir_amos, with key contributions from Yuval Tamir and @yaelam75. This would not have been possible without the great collaboration with @leeat_keren! And thanks to @WellcomeLeap ΔTissue for funding!
16/n
16/n
CISM source code is publicly available github.com/zaritskylab/...
Also, check CISM’s backbone FANMOD+, that enables efficient motifs extraction in “multi-colored” networks github.com/zaritskylab/...
Try it out!
15/n
Also, check CISM’s backbone FANMOD+, that enables efficient motifs extraction in “multi-colored” networks github.com/zaritskylab/...
Try it out!
15/n
GitHub - zaritskylab/CISM: A two-step method to identify local interconnected cell structures associated with a disease state in single cell spatial proteomics data
A two-step method to identify local interconnected cell structures associated with a disease state in single cell spatial proteomics data - zaritskylab/CISM
github.com
May 9, 2025 at 9:08 AM
CISM source code is publicly available github.com/zaritskylab/...
Also, check CISM’s backbone FANMOD+, that enables efficient motifs extraction in “multi-colored” networks github.com/zaritskylab/...
Try it out!
15/n
Also, check CISM’s backbone FANMOD+, that enables efficient motifs extraction in “multi-colored” networks github.com/zaritskylab/...
Try it out!
15/n
Our results suggest that the local organization of a few cells in discriminative motifs are emergent properties that may define an intermediate spatial scale driving tissue function
14/n
14/n

May 9, 2025 at 9:08 AM
Our results suggest that the local organization of a few cells in discriminative motifs are emergent properties that may define an intermediate spatial scale driving tissue function
14/n
14/n
CISM’s unsupervised motif enumeration followed by supervised context-dependent selection distils the discriminative motifs from the huge and noisy landscape of all putative sub-networks of intercellular interactions and provides discrimination along with interpretability
13/n
13/n

May 9, 2025 at 9:08 AM
CISM’s unsupervised motif enumeration followed by supervised context-dependent selection distils the discriminative motifs from the huge and noisy landscape of all putative sub-networks of intercellular interactions and provides discrimination along with interpretability
13/n
13/n
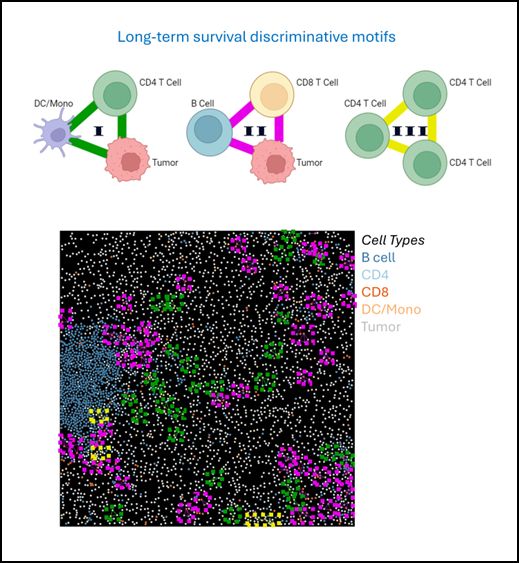
To demonstrate the general applicability of CISM for identifying local cellular structures linked to disease states, we applied it to investigate the tumor microenvironment in a human cohort of Breast Cancer (TNBC) patients, comparing short-term and long-term survivors.
12/n
12/n
May 9, 2025 at 9:08 AM
To demonstrate the general applicability of CISM for identifying local cellular structures linked to disease states, we applied it to investigate the tumor microenvironment in a human cohort of Breast Cancer (TNBC) patients, comparing short-term and long-term survivors.
12/n
12/n
The same motifs can be differentially analyzed according to different context-dependent selection of the discriminative motifs. We demonstrate this by investigating the immune microenvironment of NP versus PN (metastatic lymph nodes that did not develop distant metastases)
11/n
11/n

May 9, 2025 at 9:08 AM
The same motifs can be differentially analyzed according to different context-dependent selection of the discriminative motifs. We demonstrate this by investigating the immune microenvironment of NP versus PN (metastatic lymph nodes that did not develop distant metastases)
11/n
11/n
Spatial interpretation of motifs localization patterns reveals an association between (local) motifs to (global) tissue compartments highlighting the potential contribution of CISM to multi-scale analysis and interpretation
10/n
10/n

May 9, 2025 at 9:08 AM
Spatial interpretation of motifs localization patterns reveals an association between (local) motifs to (global) tissue compartments highlighting the potential contribution of CISM to multi-scale analysis and interpretation
10/n
10/n
The spatial arrangement of cell types within the discriminative motifs contributed to disease state classification, indicating that the specific intra-motif cell-cell interactions are sensitive markers for disease state beyond their cell type composition
9/n
9/n

May 9, 2025 at 9:08 AM
The spatial arrangement of cell types within the discriminative motifs contributed to disease state classification, indicating that the specific intra-motif cell-cell interactions are sensitive markers for disease state beyond their cell type composition
9/n
9/n
Exploring the landscape of the discriminative motifs-induced cell distribution and motifs-induced pairwise cell-cell interactions revealed differential composition of cell type and cell-cell interactions
8/n
8/n

May 9, 2025 at 9:08 AM
Exploring the landscape of the discriminative motifs-induced cell distribution and motifs-induced pairwise cell-cell interactions revealed differential composition of cell type and cell-cell interactions
8/n
8/n
Classifying NN vs. NP disease states using discriminative four-cell motifs outperformed other methods, suggesting that these motifs can act as multicellular signatures of disease
7/n
7/n

May 9, 2025 at 9:08 AM
Classifying NN vs. NP disease states using discriminative four-cell motifs outperformed other methods, suggesting that these motifs can act as multicellular signatures of disease
7/n
7/n

