https://andrewkennard.github.io
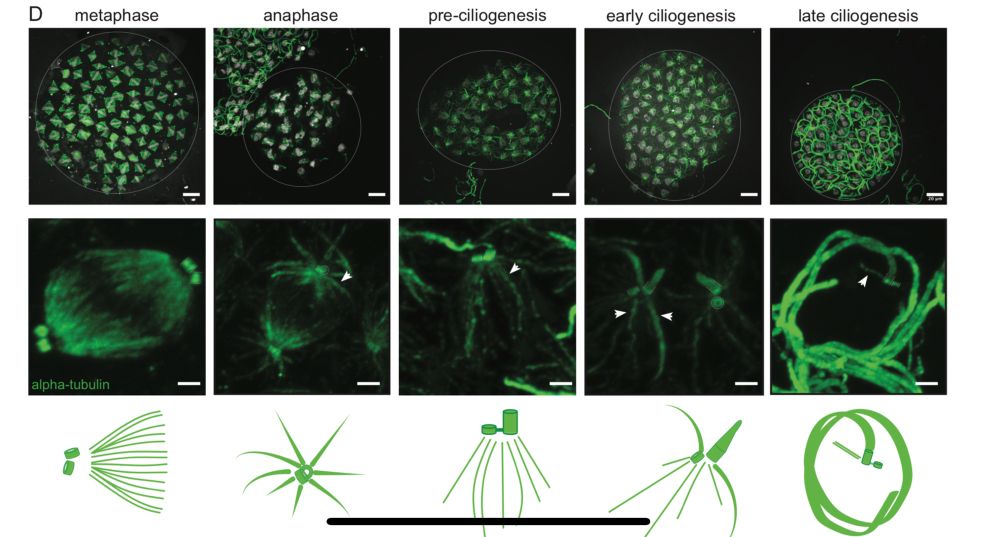


2) Naegleria builds mitotic spindles and flagella with different tubulins. So Naegleria makes temporally, functionally, and genetically distinct microtubule networks. (6/14)

2) Naegleria builds mitotic spindles and flagella with different tubulins. So Naegleria makes temporally, functionally, and genetically distinct microtubule networks. (6/14)
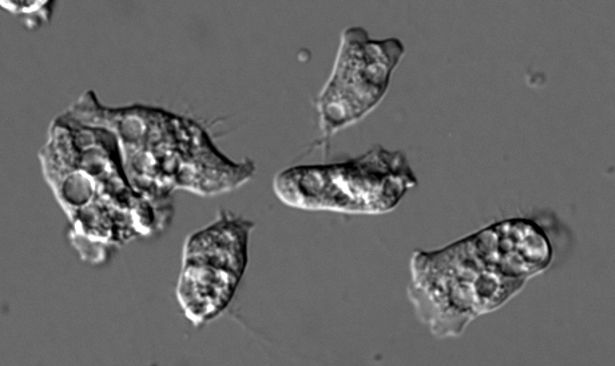
Our star is Naegleria gruberi, an amoeba separated from us by about 1.5 billion years of evolution. Naegleria is a fantastic model for microtubule regulation: (5/14)
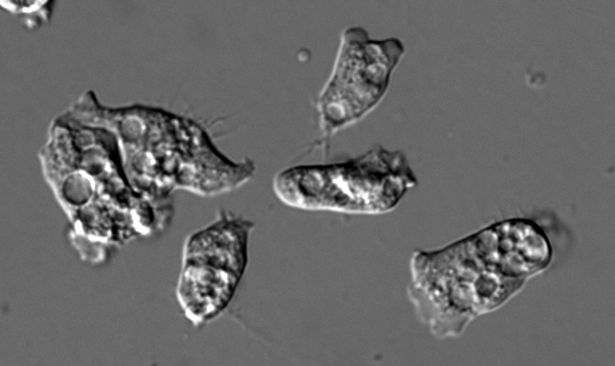
Our star is Naegleria gruberi, an amoeba separated from us by about 1.5 billion years of evolution. Naegleria is a fantastic model for microtubule regulation: (5/14)