
We had a huge task—picking bacterial stocks from >200 96-well plates and re-arranging them—and the lab's anaerobic liquid handler was broken without the funds to repair. 🧵

We had a huge task—picking bacterial stocks from >200 96-well plates and re-arranging them—and the lab's anaerobic liquid handler was broken without the funds to repair. 🧵
We identified new genes involved in aromatic lactic acid production, an important set of signals to the developing infant, and connected production of these small molecules to growth of bifidobacteria.
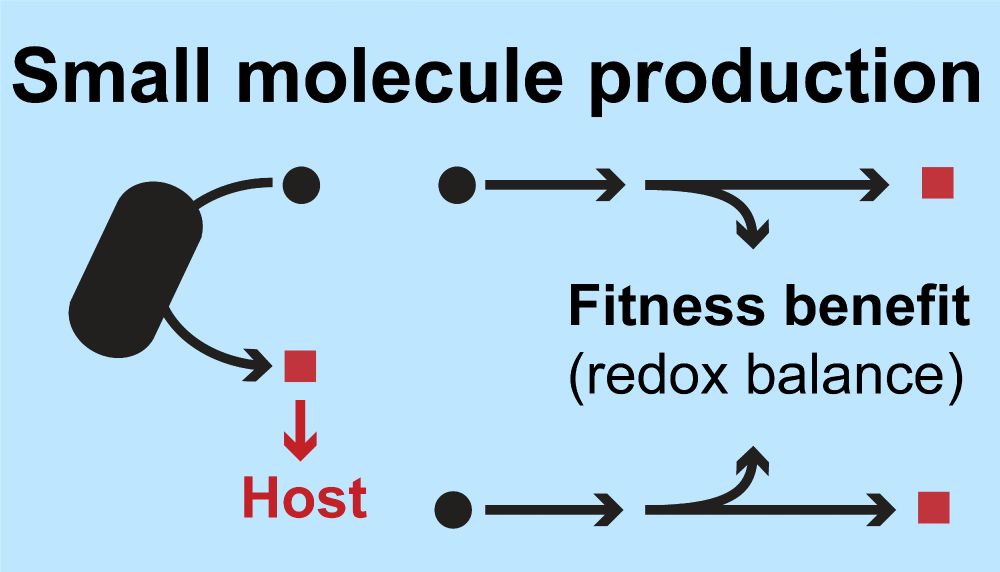
We identified new genes involved in aromatic lactic acid production, an important set of signals to the developing infant, and connected production of these small molecules to growth of bifidobacteria.
By referencing our metabolic reconstruction, we were able to paint a better picture of growth in the complex environment of the host guts. We learned about persistence in adults and the timing of colonization in infants.
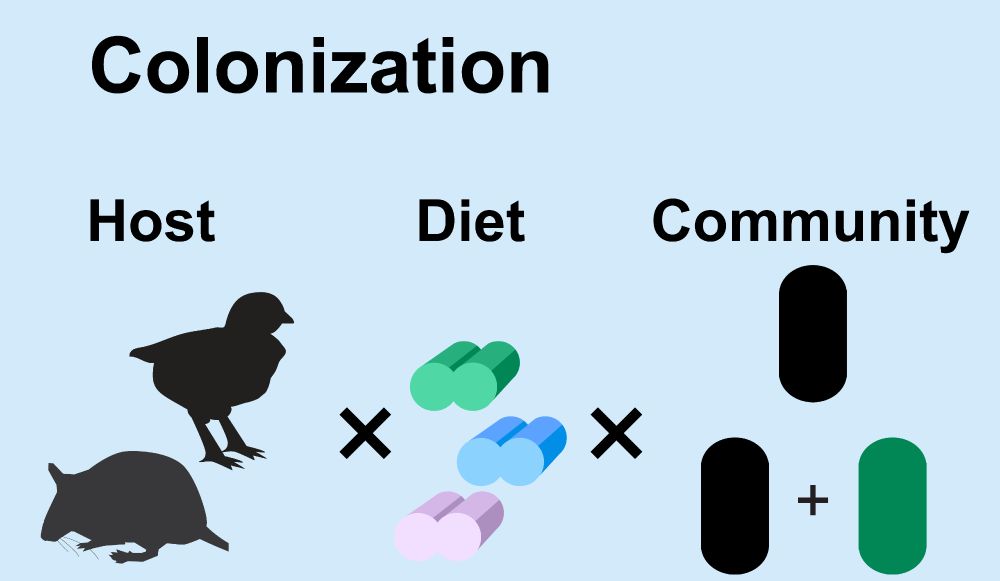
By referencing our metabolic reconstruction, we were able to paint a better picture of growth in the complex environment of the host guts. We learned about persistence in adults and the timing of colonization in infants.
With experimentally supported metabolic pathways in hand, we then tackled growth in more complex environments like the gut.
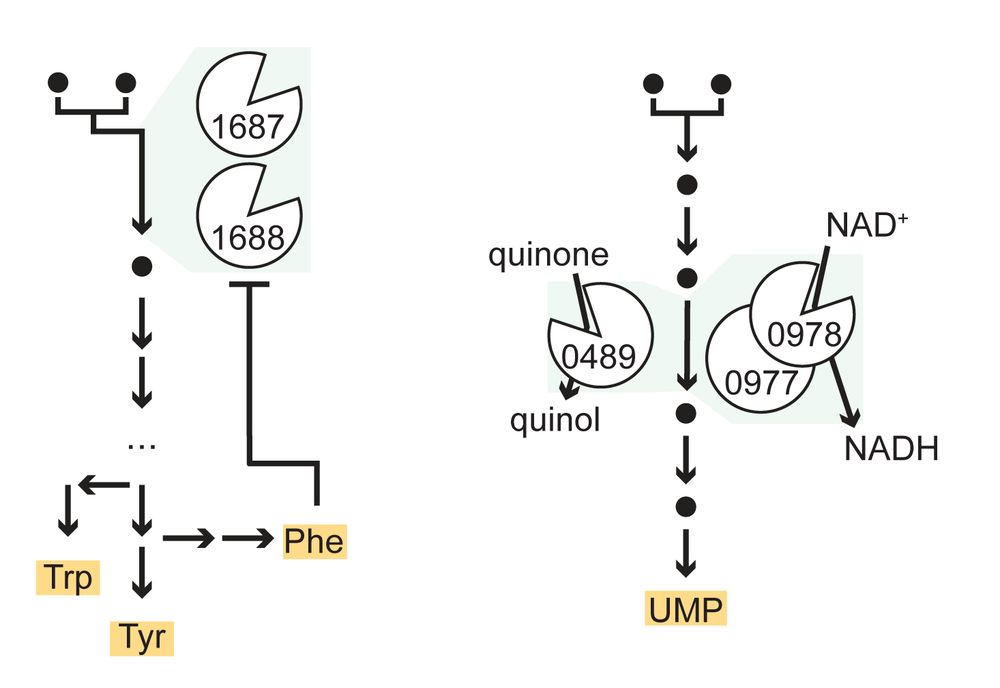
With experimentally supported metabolic pathways in hand, we then tackled growth in more complex environments like the gut.
We used this pool to conduct a chemical-genomic screen and converted the transposon pool into an ordered mutant collection.
We then tackled multiple aspects of bifidobacterial biology with these resources.
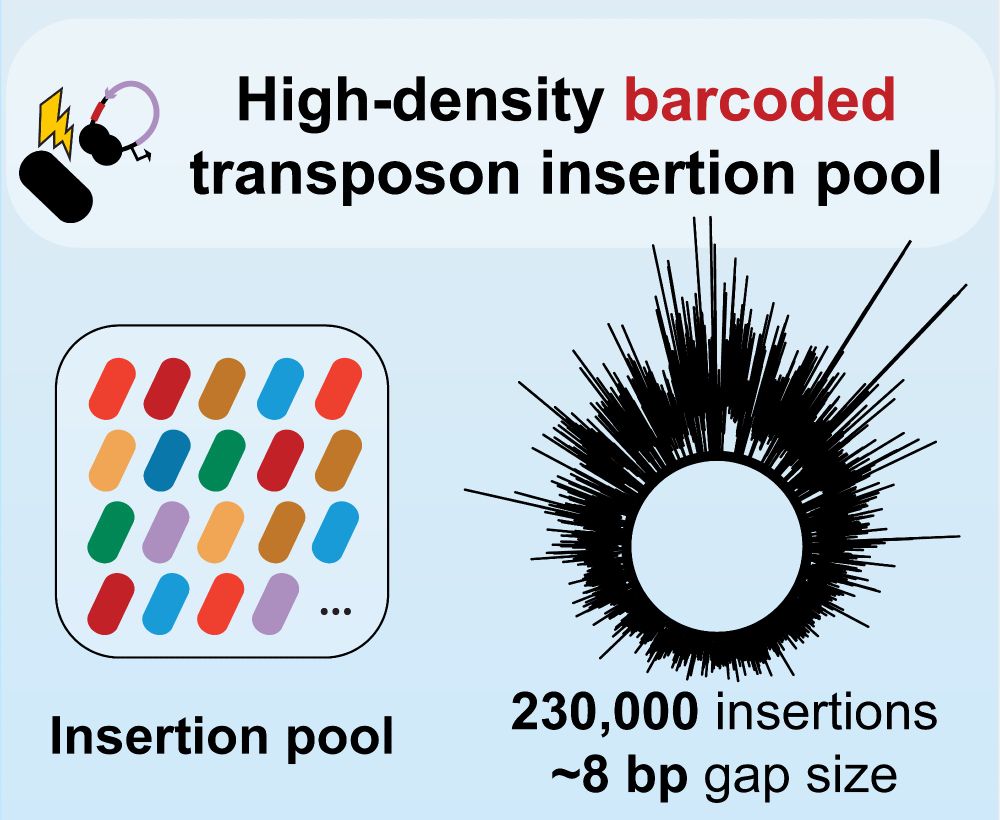
We used this pool to conduct a chemical-genomic screen and converted the transposon pool into an ordered mutant collection.
We then tackled multiple aspects of bifidobacterial biology with these resources.
A new publication, describing genome-scale resources in bifidobacteria, is now available online: doi.org/10.1016/j.ce...
#microbiome #microsky

A new publication, describing genome-scale resources in bifidobacteria, is now available online: doi.org/10.1016/j.ce...
#microbiome #microsky
By referencing our metabolic reconstruction, we were able to paint a better picture of growth in the complex environment of the host guts. We learned about persistence in adults and the timing of colonization in infants.


By referencing our metabolic reconstruction, we were able to paint a better picture of growth in the complex environment of the host guts. We learned about persistence in adults and the timing of colonization in infants.
With experimentally supported metabolic pathways in hand, we then tackled growth in more complex environments like the gut.


With experimentally supported metabolic pathways in hand, we then tackled growth in more complex environments like the gut.
We used this pool to conduct a chemical-genomic screen and converted the transposon pool into an ordered mutant collection.
We then tackled multiple aspects of bifidobacterial biology with these resources.



We used this pool to conduct a chemical-genomic screen and converted the transposon pool into an ordered mutant collection.
We then tackled multiple aspects of bifidobacterial biology with these resources.
How did we do this?

How did we do this?

