
Microscopy of bacteria and their viruses.
Postdoc at Yale University @paulturnerlab.bsky.social. Phage researcher, biotech enthusiast.
Research Facilitator at Optical Microscopy and Imaging in Biomedical Sciences (MBL, Woods H


Max Z projections from just widefield fluorescence images! 💙🔬🤩
#microscopy with Henry Arenas-Castro @jenncoughlan.bsky.social lab


Max Z projections from just widefield fluorescence images! 💙🔬🤩
#microscopy with Henry Arenas-Castro @jenncoughlan.bsky.social lab

@asm.org ASM Studio
#ASMicrobe #ASMicrobe2025
asm.org/articles/sal...

@asm.org ASM Studio
#ASMicrobe #ASMicrobe2025
asm.org/articles/sal...

Wrote a piece describing our recent paper @paulturnerlab.bsky.social to a high-school educated audience. #phagesky #microsky
Reach out if you want to do the same for your recent paper. It's super fun!
www.journaloflifesciences.org/archives/156...



Wrote a piece describing our recent paper @paulturnerlab.bsky.social to a high-school educated audience. #phagesky #microsky
Reach out if you want to do the same for your recent paper. It's super fun!
www.journaloflifesciences.org/archives/156...
We evolved bacteria on swim plates, against a phage that attacks through the flagellum. Motility trades off AND trades up(!!) with phage-resistance!
www.biorxiv.org/content/10.1...
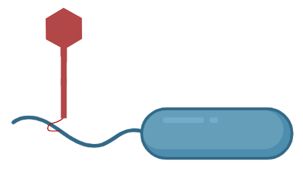
We evolved bacteria on swim plates, against a phage that attacks through the flagellum. Motility trades off AND trades up(!!) with phage-resistance!
www.biorxiv.org/content/10.1...
I asked the tattoo-artist to draw the podovirus where the genome is cousin's wedding logo.
#phagesky

I asked the tattoo-artist to draw the podovirus where the genome is cousin's wedding logo.
#phagesky

The time a virus stays in view under the microscope (trajectory duration) reflects how long it closely interacts with host cells (Dwell Time). 🕒 We observed massive variability in these interactions—each virus behaves differently!

The time a virus stays in view under the microscope (trajectory duration) reflects how long it closely interacts with host cells (Dwell Time). 🕒 We observed massive variability in these interactions—each virus behaves differently!




The paper was posted online today:
www.pnas.org/doi/10.1073/...
#phagesky #microsky
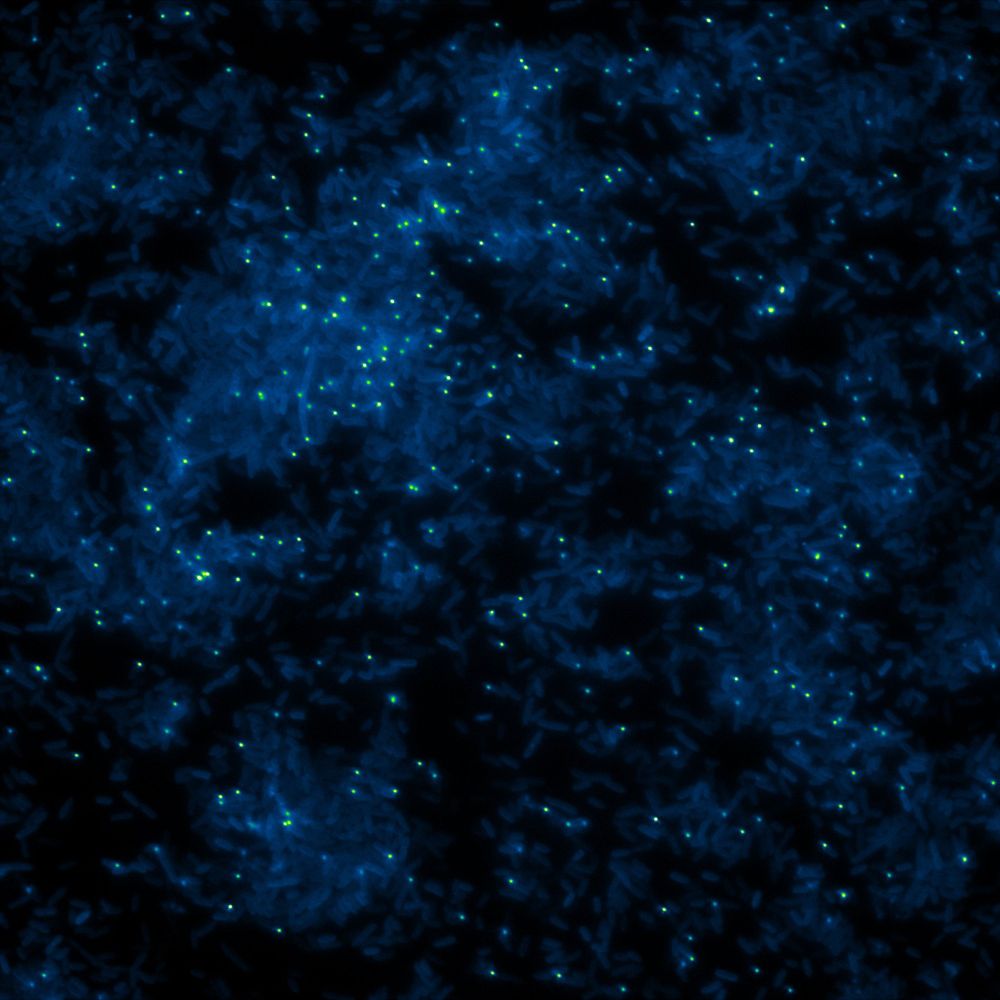
The paper was posted online today:
www.pnas.org/doi/10.1073/...
#phagesky #microsky


