
Amir Mitchell
@amitchell.bsky.social
Systems biologist trying to disentangle host-drug-microbiome interactions | PI at UMass medical school | trying to keep the BS to a minimum (not always successful)
https://mitchell-lab.umassmed.edu
https://mitchell-lab.umassmed.edu
The inherent stochasticity of biological systems 😉
January 18, 2025 at 7:46 PM
The inherent stochasticity of biological systems 😉
Amazing book, shaped my worldview on Science. Kuhn’s genius was also in claiming that “paradigm shifts” are not purely logical, but are tainted by social & psychological factors. He claimed that established scientists are often conservatives resisting such shifts.
December 28, 2024 at 3:02 AM
Amazing book, shaped my worldview on Science. Kuhn’s genius was also in claiming that “paradigm shifts” are not purely logical, but are tainted by social & psychological factors. He claimed that established scientists are often conservatives resisting such shifts.
It was all code written in Matlab
December 14, 2024 at 7:50 PM
It was all code written in Matlab
Sure, email me I can attach a copy
December 14, 2024 at 4:42 PM
Sure, email me I can attach a copy
The drug-drug similarity networks are based on how these drugs impact a library of E. coli mutants (profiles of resistance or sensitivity). We in fact show the chemical similarity fails to capture many of these similarities.
December 14, 2024 at 4:41 PM
The drug-drug similarity networks are based on how these drugs impact a library of E. coli mutants (profiles of resistance or sensitivity). We in fact show the chemical similarity fails to capture many of these similarities.
My only complain is that I feel a bit like Cassandra, from the greek mythology, barely anyone listens 😞

December 7, 2024 at 11:23 PM
My only complain is that I feel a bit like Cassandra, from the greek mythology, barely anyone listens 😞
There is also a lot to be said about doing a "proper" postdoc in a good lab. It opens your eyes to different research directions, lab techniques, mentoring style, institutions, and so much more!
December 4, 2024 at 9:17 PM
There is also a lot to be said about doing a "proper" postdoc in a good lab. It opens your eyes to different research directions, lab techniques, mentoring style, institutions, and so much more!
I absolutely agree!! So many of these positions end up going nowhere. They make sense only in *very* few use cases (5-10%): it's a *real* program with real legacy of successful PIs, the candidate continues on their phd research trajectory, and the candidate is scientifically mature (rare).
December 4, 2024 at 9:17 PM
I absolutely agree!! So many of these positions end up going nowhere. They make sense only in *very* few use cases (5-10%): it's a *real* program with real legacy of successful PIs, the candidate continues on their phd research trajectory, and the candidate is scientifically mature (rare).
Credit for this work goes to my former student Dr Emily Lowry. Emily literally submitted the revised version of this manuscript at the very last minute before leaving the lab 🥳 (9/9)
December 2, 2024 at 4:44 PM
Credit for this work goes to my former student Dr Emily Lowry. Emily literally submitted the revised version of this manuscript at the very last minute before leaving the lab 🥳 (9/9)
I love this work since it a wonderful demonstration of (1) Serendipity in research – it started unexpectedly when we saw an active reporter even in non-contacting colonies and (2) it shows the power of simple experiments and quantitative microscopy (8/9)
December 2, 2024 at 4:44 PM
I love this work since it a wonderful demonstration of (1) Serendipity in research – it started unexpectedly when we saw an active reporter even in non-contacting colonies and (2) it shows the power of simple experiments and quantitative microscopy (8/9)
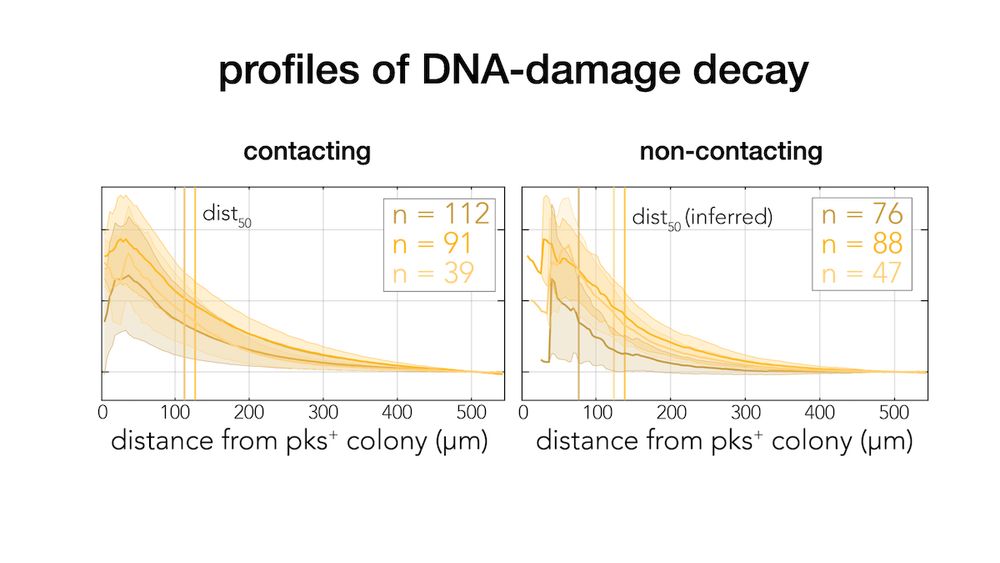
Careful quantification revealed that the damage decay curves are identical in contacting and non-contacting colonies. Therefore, direct contact is not only not required, but it doesn’t even increase the level of toxicity beyond what is expected by proximity (7/9)
December 2, 2024 at 4:44 PM
Careful quantification revealed that the damage decay curves are identical in contacting and non-contacting colonies. Therefore, direct contact is not only not required, but it doesn’t even increase the level of toxicity beyond what is expected by proximity (7/9)
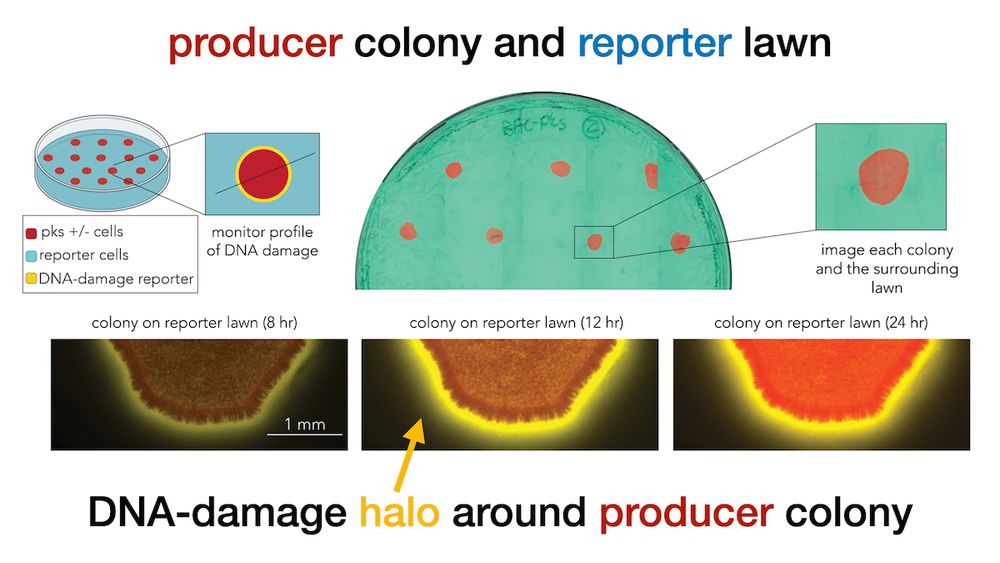
Next, we monitored colibactin damage in separated colonies. This setup allowed us not only to validate contact independence, but also to accurately quantify how DNA-damage decays across distance (6/9)
December 2, 2024 at 4:44 PM
Next, we monitored colibactin damage in separated colonies. This setup allowed us not only to validate contact independence, but also to accurately quantify how DNA-damage decays across distance (6/9)
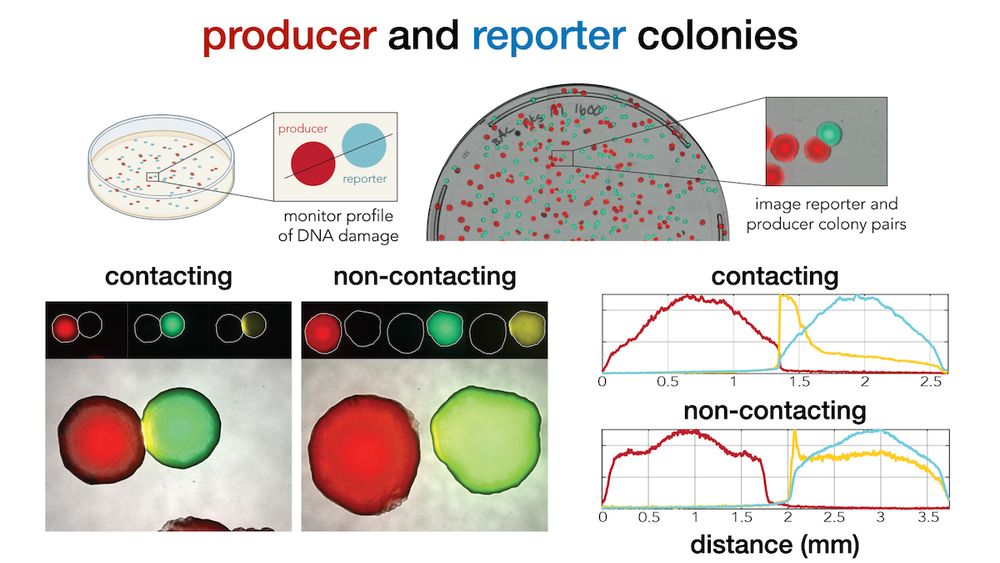
Microscopy imaging revealed that DNA damage is observed even hundreds of microns (YFP halo) away from the secreter front (mCherry) suggesting that direct cell-cell is not needed for toxicity (5/9)
December 2, 2024 at 4:44 PM
Microscopy imaging revealed that DNA damage is observed even hundreds of microns (YFP halo) away from the secreter front (mCherry) suggesting that direct cell-cell is not needed for toxicity (5/9)
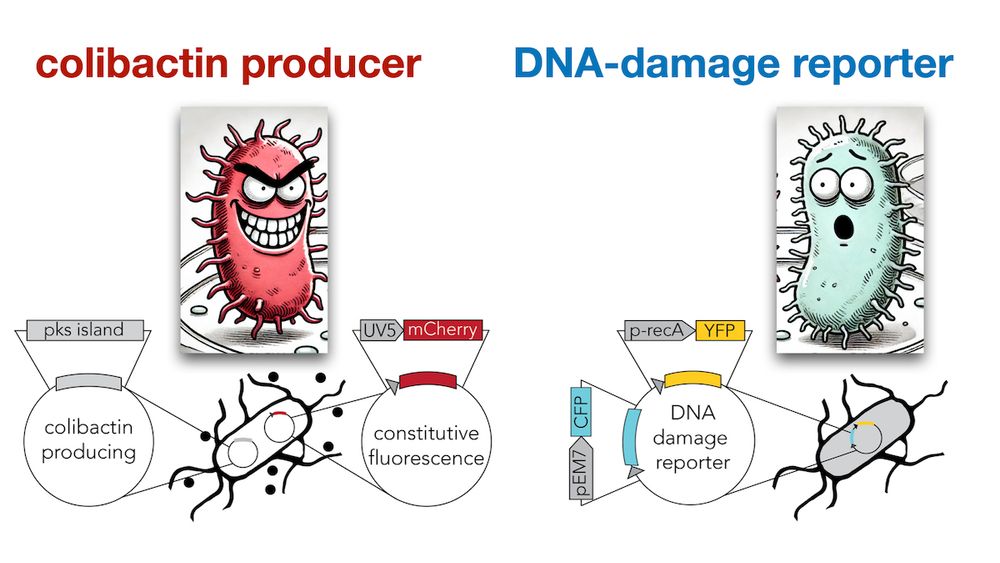
We cloned a YFP DNA-damage reporter in E. coli and tested how far colibactin induced damage “travels” across a lawn of cells (we tagged secreters and responders with constitutive mCherry and CFP to tell them apart) (4/9)
December 2, 2024 at 4:44 PM
We cloned a YFP DNA-damage reporter in E. coli and tested how far colibactin induced damage “travels” across a lawn of cells (we tagged secreters and responders with constitutive mCherry and CFP to tell them apart) (4/9)
Colibactin mode of delivery is still under debate, with common models claiming delivery requires direct cell-cell contact. While contact certainly improves toxicity (see great recent work by the Lars Vereecke lab), does it add anything beyond what is expected by proximity alone? (3/9)
December 2, 2024 at 4:44 PM
Colibactin mode of delivery is still under debate, with common models claiming delivery requires direct cell-cell contact. While contact certainly improves toxicity (see great recent work by the Lars Vereecke lab), does it add anything beyond what is expected by proximity alone? (3/9)
Colibactin is deeply studied in human cells due to its involvement in colorectal cancers, yet far less is known in the context of microbial communities. Our first study with @daganlab.bsky.social revealed self-inflicted DNA-damage in colibactin producers (2/9)
genome.cshlp.org/content/earl...
genome.cshlp.org/content/earl...
Colibactin leads to a bacteria-specific mutation pattern and self-inflicted DNA damage
An international, peer-reviewed genome sciences journal featuring outstanding original research that offers novel insights into the biology of all organisms
genome.cshlp.org
December 2, 2024 at 4:44 PM
Colibactin is deeply studied in human cells due to its involvement in colorectal cancers, yet far less is known in the context of microbial communities. Our first study with @daganlab.bsky.social revealed self-inflicted DNA-damage in colibactin producers (2/9)
genome.cshlp.org/content/earl...
genome.cshlp.org/content/earl...

