
Amir Mitchell
@amitchell.bsky.social
Systems biologist trying to disentangle host-drug-microbiome interactions | PI at UMass medical school | trying to keep the BS to a minimum (not always successful)
https://mitchell-lab.umassmed.edu
https://mitchell-lab.umassmed.edu
Interested in Antimicrobial Resistance & Quantitative Bio? Join a 3-day meeting+workshop with some of my favorite #AMR #QBio scientists this April (University of Exeter, UK). Register here: tinyurl.com/QAMR2025reg 🧪🦠
January 17, 2025 at 3:16 PM
Interested in Antimicrobial Resistance & Quantitative Bio? Join a 3-day meeting+workshop with some of my favorite #AMR #QBio scientists this April (University of Exeter, UK). Register here: tinyurl.com/QAMR2025reg 🧪🦠
My only complain is that I feel a bit like Cassandra, from the greek mythology, barely anyone listens 😞

December 7, 2024 at 11:23 PM
My only complain is that I feel a bit like Cassandra, from the greek mythology, barely anyone listens 😞
The ability of LMMs (chatGPT here) to *assist* in generating useful code for biologists a mind-blowing power multiplier 🤯! My advice to biologists at all career stages: Learn to code, its a superpower 🦸♀️🦹♂️ (simple example, counting fluorescent colonies isolated from mouse gut microbiome) 🧪🦠

December 7, 2024 at 11:04 PM
The ability of LMMs (chatGPT here) to *assist* in generating useful code for biologists a mind-blowing power multiplier 🤯! My advice to biologists at all career stages: Learn to code, its a superpower 🦸♀️🦹♂️ (simple example, counting fluorescent colonies isolated from mouse gut microbiome) 🧪🦠
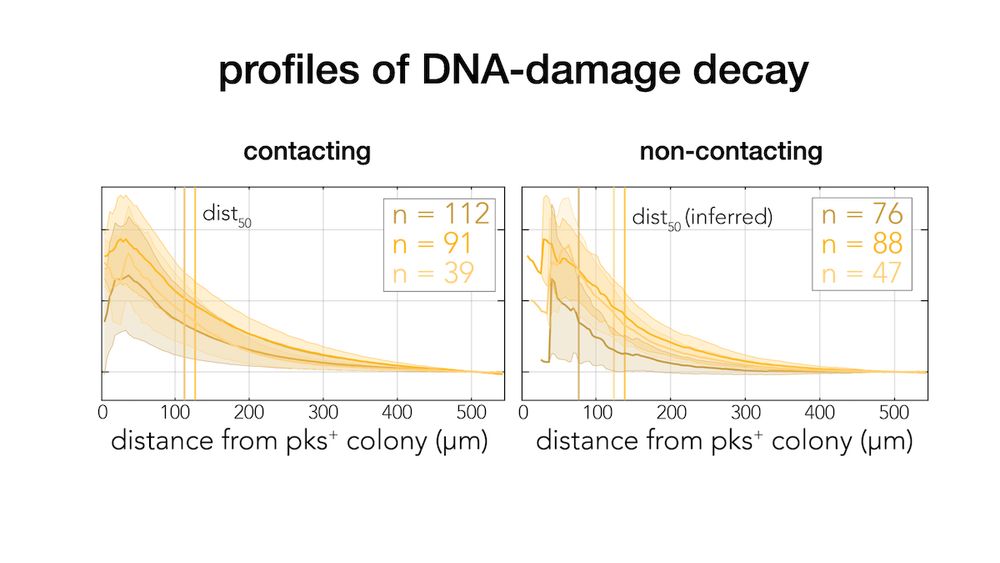
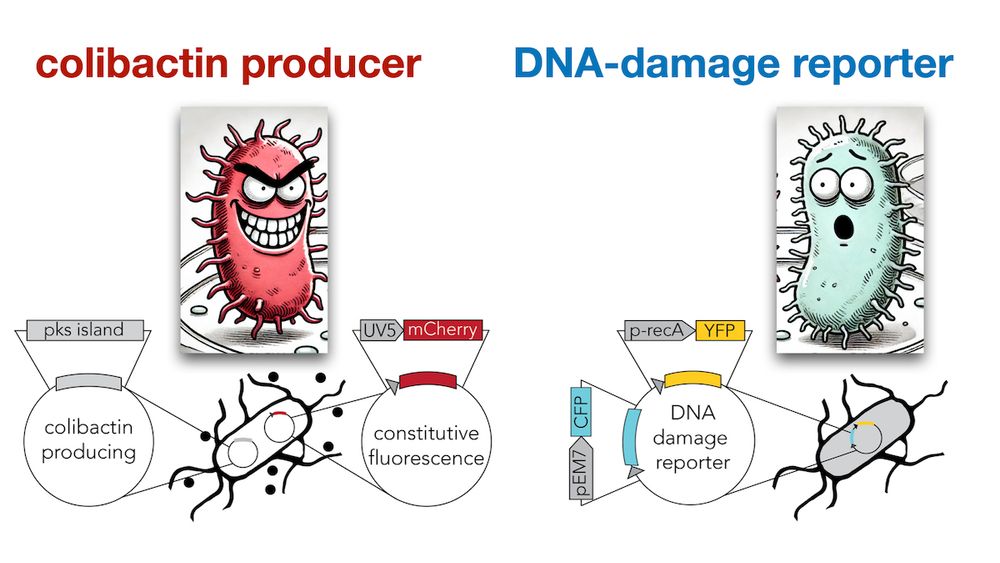
Careful quantification revealed that the damage decay curves are identical in contacting and non-contacting colonies. Therefore, direct contact is not only not required, but it doesn’t even increase the level of toxicity beyond what is expected by proximity (7/9)
December 2, 2024 at 4:44 PM
Careful quantification revealed that the damage decay curves are identical in contacting and non-contacting colonies. Therefore, direct contact is not only not required, but it doesn’t even increase the level of toxicity beyond what is expected by proximity (7/9)
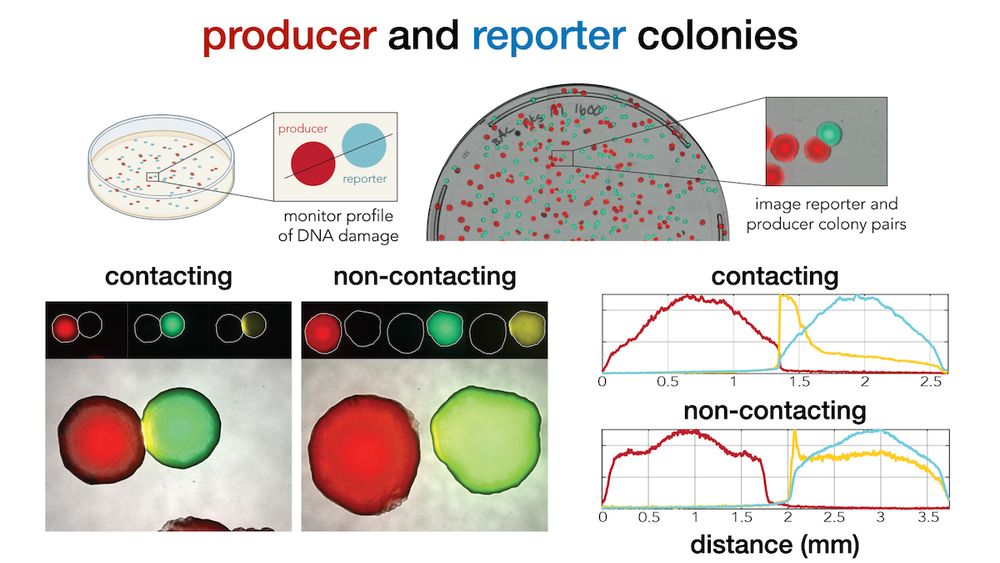
Next, we monitored colibactin damage in separated colonies. This setup allowed us not only to validate contact independence, but also to accurately quantify how DNA-damage decays across distance (6/9)
December 2, 2024 at 4:44 PM
Next, we monitored colibactin damage in separated colonies. This setup allowed us not only to validate contact independence, but also to accurately quantify how DNA-damage decays across distance (6/9)
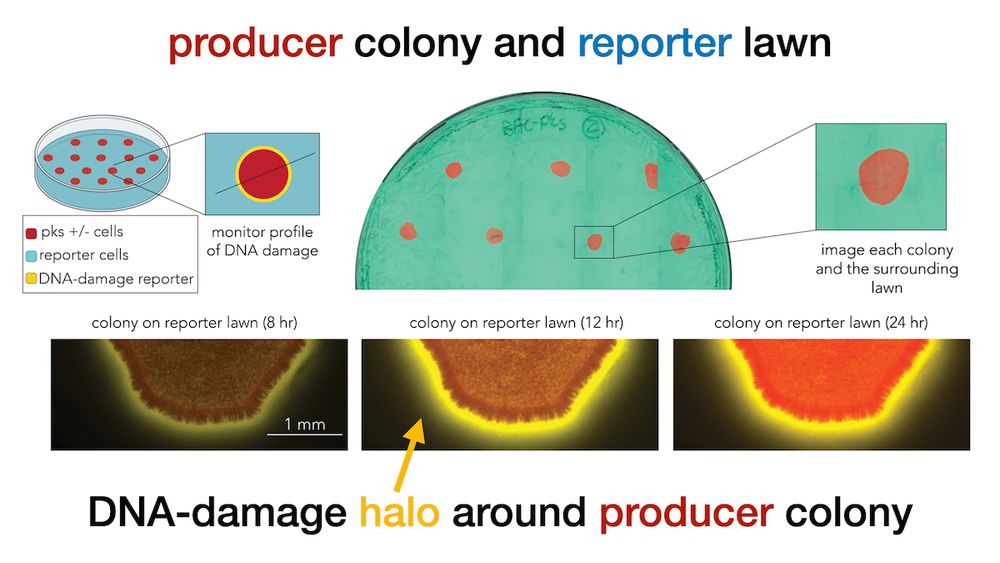
Microscopy imaging revealed that DNA damage is observed even hundreds of microns (YFP halo) away from the secreter front (mCherry) suggesting that direct cell-cell is not needed for toxicity (5/9)
December 2, 2024 at 4:44 PM
Microscopy imaging revealed that DNA damage is observed even hundreds of microns (YFP halo) away from the secreter front (mCherry) suggesting that direct cell-cell is not needed for toxicity (5/9)
We cloned a YFP DNA-damage reporter in E. coli and tested how far colibactin induced damage “travels” across a lawn of cells (we tagged secreters and responders with constitutive mCherry and CFP to tell them apart) (4/9)
December 2, 2024 at 4:44 PM
We cloned a YFP DNA-damage reporter in E. coli and tested how far colibactin induced damage “travels” across a lawn of cells (we tagged secreters and responders with constitutive mCherry and CFP to tell them apart) (4/9)
Our second paper on the bacterial toxin colibactin is now out on mBio, this time we critically evaluated the claim that colibactin toxicity necessitates cell-cell contact 🧪🦠🧵(1/9)
journals.asm.org/doi/10.1128/...
journals.asm.org/doi/10.1128/...
December 2, 2024 at 4:44 PM
Our second paper on the bacterial toxin colibactin is now out on mBio, this time we critically evaluated the claim that colibactin toxicity necessitates cell-cell contact 🧪🦠🧵(1/9)
journals.asm.org/doi/10.1128/...
journals.asm.org/doi/10.1128/...
Can’t believe we unlocked this level of the #AI hype - huge adds for @AnthropicAI in airports (Boston & Atlanta). Wonder what % of ppl even know what’s the product (<0.1% probably)

November 23, 2024 at 6:14 AM
Can’t believe we unlocked this level of the #AI hype - huge adds for @AnthropicAI in airports (Boston & Atlanta). Wonder what % of ppl even know what’s the product (<0.1% probably)
As a validation of our hypothesis on self-inflicted damage, we genetically engineered a fluorescent reporter of DNA damage and found its basal activity is indeed noticeably increased in colibactin producing cells (either engineered or natural strains)

November 23, 2024 at 7:06 AM
As a validation of our hypothesis on self-inflicted damage, we genetically engineered a fluorescent reporter of DNA damage and found its basal activity is indeed noticeably increased in colibactin producing cells (either engineered or natural strains)
Exploiting the mut signature we discovered, we scanned 10K E. coli genomes in search for compatible genomic scars. Surprisingly, we found that such scars are enriched in strains that produce colibactin. This indicates that producers cannot(!) avoid some self-inflicted damage

November 23, 2024 at 6:58 AM
Exploiting the mut signature we discovered, we scanned 10K E. coli genomes in search for compatible genomic scars. Surprisingly, we found that such scars are enriched in strains that produce colibactin. This indicates that producers cannot(!) avoid some self-inflicted damage
Yet, although in bacteria colibactin favored binding A/T rich oligos, just like in humans, colibactin damage led to to a bacteria-specific mutation profile – mostly T->A (rather than T->C)

November 23, 2024 at 6:50 AM
Yet, although in bacteria colibactin favored binding A/T rich oligos, just like in humans, colibactin damage led to to a bacteria-specific mutation profile – mostly T->A (rather than T->C)
We then tested the genomic changes colibactin induces with a mutation accumulation experiment (in a 100 parallel replicates). We repetitively exposed bacteria to colibactin producing cells and then sequenced their genomes (#WGS) to infer the mutational landscape

November 23, 2024 at 6:34 AM
We then tested the genomic changes colibactin induces with a mutation accumulation experiment (in a 100 parallel replicates). We repetitively exposed bacteria to colibactin producing cells and then sequenced their genomes (#WGS) to infer the mutational landscape
We first systematically mapped all genes modulating colibactin toxicity in targeted bacteria with a loss-of-function genetic screen and found that multiple pathways, beyond nucleotide excision repair, influence colibactin sensitivity (HR, nuc synthesis, stringent)

November 23, 2024 at 6:28 AM
We first systematically mapped all genes modulating colibactin toxicity in targeted bacteria with a loss-of-function genetic screen and found that multiple pathways, beyond nucleotide excision repair, influence colibactin sensitivity (HR, nuc synthesis, stringent)
Past works showed that colibactin cross-links DNA and suggested that 5-10% of colorectal cancers may arise from colibactin producing E. coli in the gut #microbiome. We set out to study how colibactin damages bacteria that may encounter it in the gut niche.

November 23, 2024 at 6:21 AM
Past works showed that colibactin cross-links DNA and suggested that 5-10% of colorectal cancers may arise from colibactin producing E. coli in the gut #microbiome. We set out to study how colibactin damages bacteria that may encounter it in the gut niche.
We found that the bacterial #toxin colibactin acts as a double-edged sword and inflicts self-damage (evident in hundreds of genomes). Our recent collaboration with @DaganLab is now out on @genomeresearch
https://genome.cshlp.org/content/34/8/1154.full
https://genome.cshlp.org/content/34/8/1154.full

November 23, 2024 at 6:14 AM
We found that the bacterial #toxin colibactin acts as a double-edged sword and inflicts self-damage (evident in hundreds of genomes). Our recent collaboration with @DaganLab is now out on @genomeresearch
https://genome.cshlp.org/content/34/8/1154.full
https://genome.cshlp.org/content/34/8/1154.full
Immensely proud of the latest graduate from our lab @emily_lowry23 🎊🎉🥳. Dr Lowry’s second paper will be published *very* soon … (and hopefully her third paper shortly after that 🤞)

November 23, 2024 at 6:14 AM
Immensely proud of the latest graduate from our lab @emily_lowry23 🎊🎉🥳. Dr Lowry’s second paper will be published *very* soon … (and hopefully her third paper shortly after that 🤞)
What makes #phd s so hard & depressing (yet also so rewarding), a thread started by @ThePhDPlace made me reflect about my own experience and my students’ journeys. Hoping this self-reflection thread will be useful for students (and PIs too!) 1/9

November 23, 2024 at 6:14 AM
What makes #phd s so hard & depressing (yet also so rewarding), a thread started by @ThePhDPlace made me reflect about my own experience and my students’ journeys. Hoping this self-reflection thread will be useful for students (and PIs too!) 1/9
Results are in, mostly saying free (academic) labor is reasonable (seriously?🤦♂️). Going forward, I'll review 2 of every 3 such grants I'm asked about 🫡

November 23, 2024 at 6:28 AM
Results are in, mostly saying free (academic) labor is reasonable (seriously?🤦♂️). Going forward, I'll review 2 of every 3 such grants I'm asked about 🫡
Signing off a very exciting week at the co-chair of the @GordonConf on Drug Resistance. Hard, yet totally worth it! Lots to share with chairs-to-be (thread on Monday)

November 23, 2024 at 6:21 AM
Signing off a very exciting week at the co-chair of the @GordonConf on Drug Resistance. Hard, yet totally worth it! Lots to share with chairs-to-be (thread on Monday)
First time I'm attending a conf with 2/3 of my lab (@GordonConf / drug resistance). Strongly recommend PIs to do that, costly, *but* a really great bonding experience.

November 23, 2024 at 6:28 AM
First time I'm attending a conf with 2/3 of my lab (@GordonConf / drug resistance). Strongly recommend PIs to do that, costly, *but* a really great bonding experience.
Deadline for @GordonConf on Drug Resistance is fast approaching (5/26). Apply by 5/21 for posters and talks. We’ll greatly appreciate help spreading the word 🙏🏼
https://www.grc.org/drug-resistance-conference/2024/
https://www.grc.org/drug-resistance-conference/2024/

November 23, 2024 at 6:35 AM
Deadline for @GordonConf on Drug Resistance is fast approaching (5/26). Apply by 5/21 for posters and talks. We’ll greatly appreciate help spreading the word 🙏🏼
https://www.grc.org/drug-resistance-conference/2024/
https://www.grc.org/drug-resistance-conference/2024/
Many people are behind this success, but this is primarily the triumph 🏆 of @MarianaNoto who confronted a challenge that seemed truly intractable (study the MOA of almost 200 drugs) (18/19)

November 23, 2024 at 9:11 AM
Many people are behind this success, but this is primarily the triumph 🏆 of @MarianaNoto who confronted a challenge that seemed truly intractable (study the MOA of almost 200 drugs) (18/19)
Even more exciting, resistant IF2 mutants also resisted two additional non-ABX drugs that were network neighbors of the drug used for evolution (pink). This means our network can uncover shared MOA even if the MOA is unknown! (16/19)

November 23, 2024 at 8:55 AM
Even more exciting, resistant IF2 mutants also resisted two additional non-ABX drugs that were network neighbors of the drug used for evolution (pink). This means our network can uncover shared MOA even if the MOA is unknown! (16/19)
Lastly, we tested our conclusions by evolving resistance against 3 non-ABX. The results supported our predictions: resistance to one non-ABX drug increased resistance against multiple ABX (by mutating regulators of pumps) (14/19)

November 23, 2024 at 8:40 AM
Lastly, we tested our conclusions by evolving resistance against 3 non-ABX. The results supported our predictions: resistance to one non-ABX drug increased resistance against multiple ABX (by mutating regulators of pumps) (14/19)

