www.nature.com/articles/s41...
www.nature.com/articles/s41...
✨ Now with LogMD-based trajectory visualization.
🔗 Demo: rcsb.ai/ff9c2b1ee8
Feedback & collabs welcome! 🙌
🔗: GitHub: github.com/yehlincho/Bo...
🔗: Colab: colab.research.google.com/github/yehli...
@sokrypton.org @martinpacesa.bsky.social
✨ Now with LogMD-based trajectory visualization.
🔗 Demo: rcsb.ai/ff9c2b1ee8
Feedback & collabs welcome! 🙌
🔗: GitHub: github.com/yehlincho/Bo...
🔗: Colab: colab.research.google.com/github/yehli...
@sokrypton.org @martinpacesa.bsky.social
www.nature.com/articles/s41...
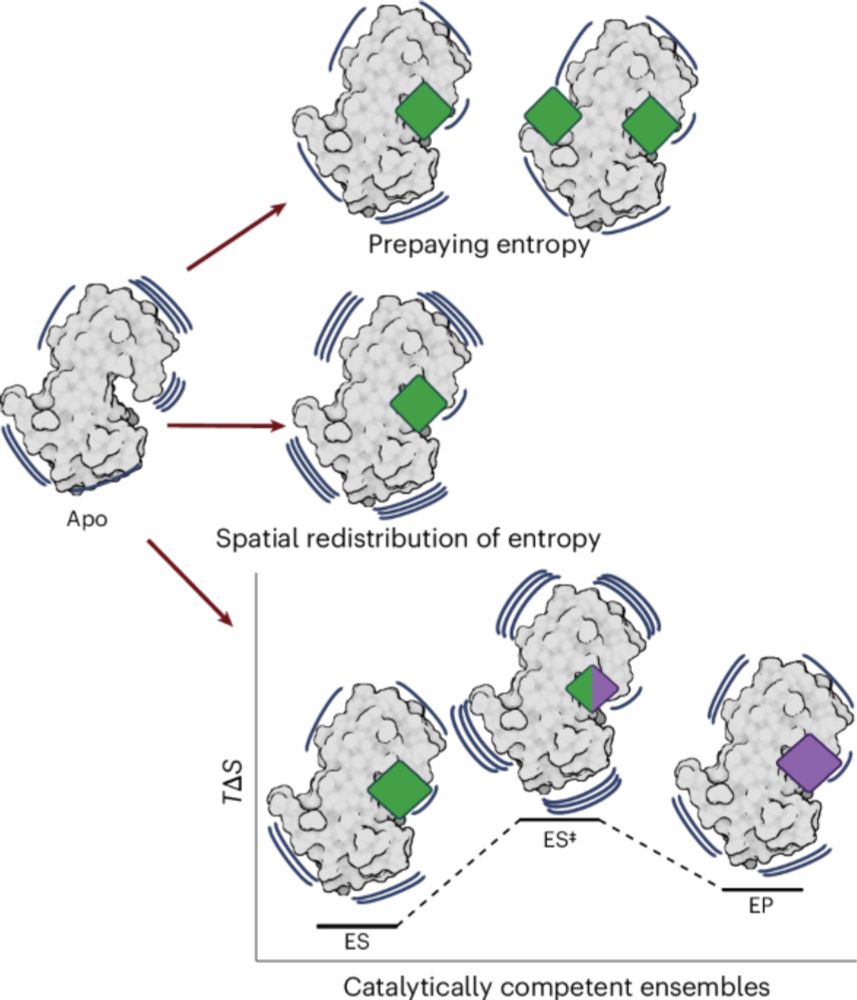
www.biorxiv.org/content/10.1...

So far, we've not had much luck in raising funds... 💰 In this relatively popular area, so I feel like I'm doing something wrong 😅
So far, we've not had much luck in raising funds... 💰 In this relatively popular area, so I feel like I'm doing something wrong 😅
@martinpacesa.bsky.social, @Zhidian Zhang, @Bruno E. Correia, and @sokrypton.org
🧬 Code will be released in a couple weeks
@martinpacesa.bsky.social, @Zhidian Zhang, @Bruno E. Correia, and @sokrypton.org
🧬 Code will be released in a couple weeks
Work by @yehlincho.bsky.social et al.
www.biorxiv.org/content/10.1...

Work by @yehlincho.bsky.social et al.
www.biorxiv.org/content/10.1...
peppr.vant.ai
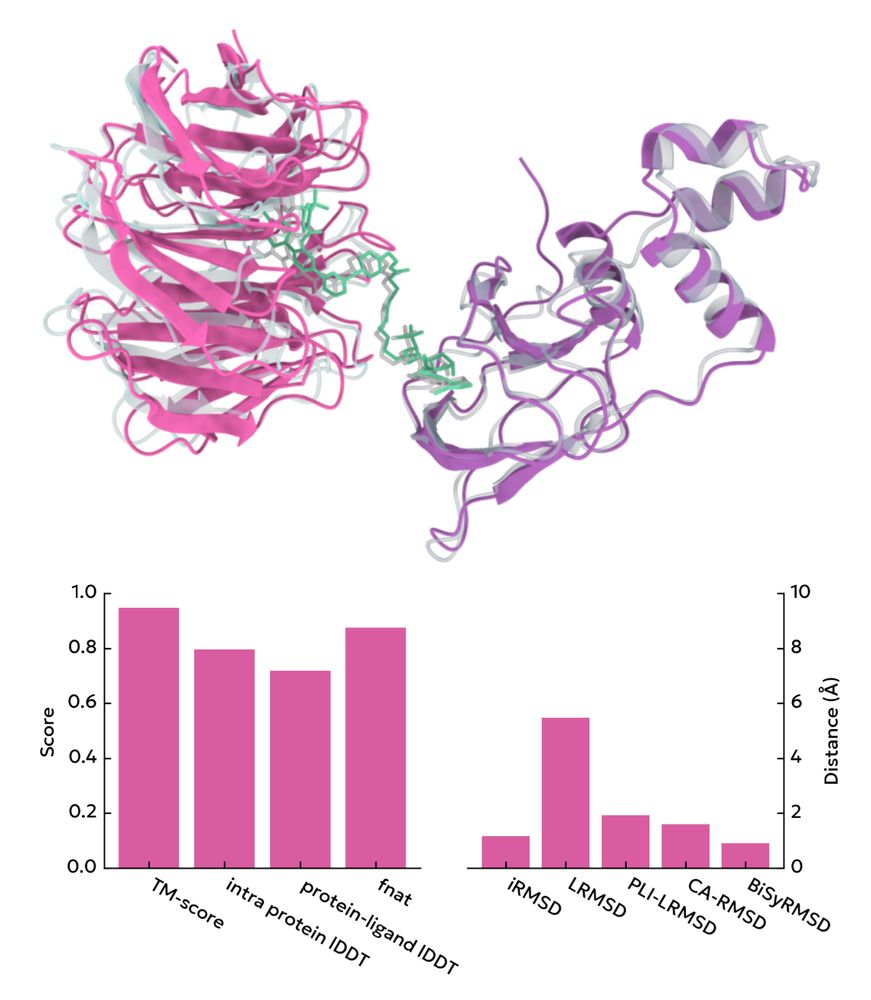
peppr.vant.ai
Here’s a quick thread of my personal favorites from the cutting edge of protein ML, dynamics, and design:
Here’s a quick thread of my personal favorites from the cutting edge of protein ML, dynamics, and design:
If you’re around, hit me up — would love to catch up with familiar faces and meet new ones! #KSMLStructure25

If you’re around, hit me up — would love to catch up with familiar faces and meet new ones! #KSMLStructure25

