I’m looking forward to building ML models that better reflect how molecules & cells look in real life 🔬
And for 2025 Fellow @alissahummer.com it is a homecoming!
Our 2025 Fellows' stories and our Science Leadership Program exemplify our mission.
schmidtsciencefellows.org/news/our-202...

I’m looking forward to building ML models that better reflect how molecules & cells look in real life 🔬
www.nature.com/natcomputsci...

www.nature.com/natcomputsci...

www.nature.com/natcomputsci...
and colleagues present Graphinity, a method to predict change in antibody-antigen binding affinity (∆∆G). Also featuring synthetic datasets of ~1 million FoldX-generated and >20,000 Rosetta Flex ddG-generated ∆∆G values!
www.nature.com/articles/s43...
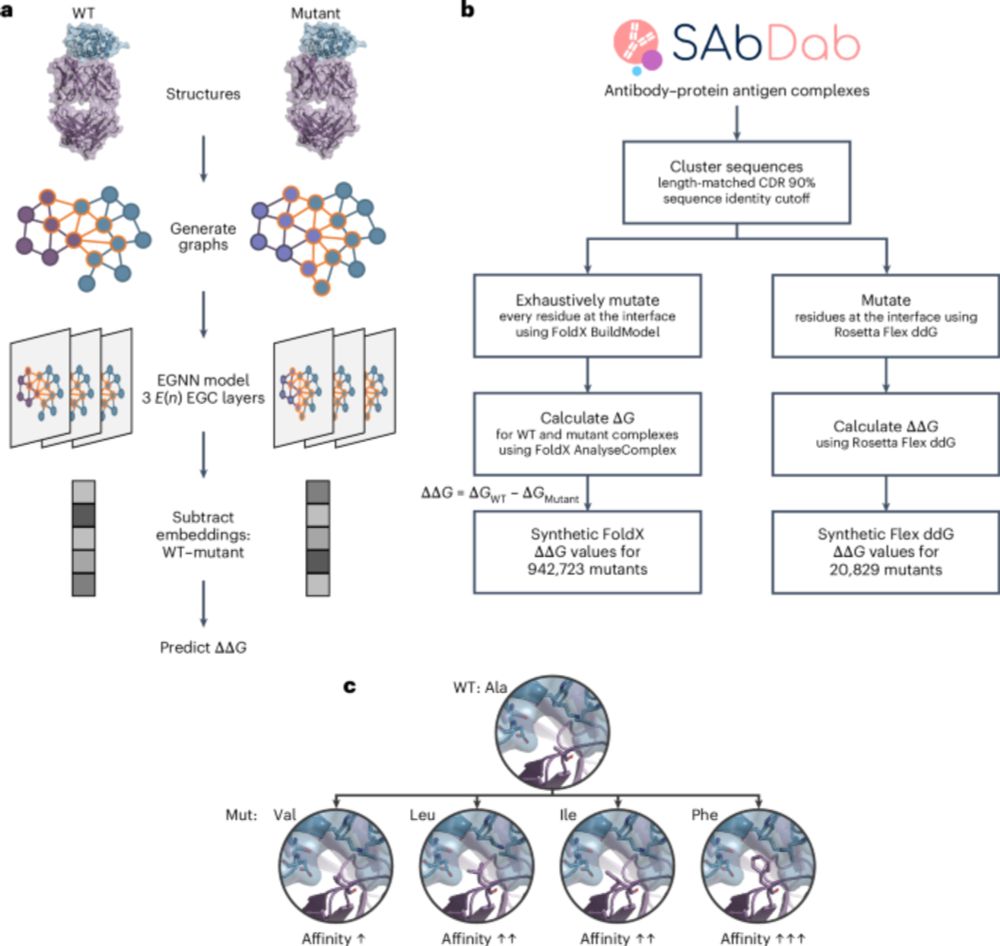
and colleagues present Graphinity, a method to predict change in antibody-antigen binding affinity (∆∆G). Also featuring synthetic datasets of ~1 million FoldX-generated and >20,000 Rosetta Flex ddG-generated ∆∆G values!
www.nature.com/articles/s43...
and colleagues present Graphinity, a method to predict change in antibody-antigen binding affinity (∆∆G). Also featuring synthetic datasets of ~1 million FoldX-generated and >20,000 Rosetta Flex ddG-generated ∆∆G values!
www.nature.com/articles/s43...


RNA modeling remains challenging for deep learning, esp. in the absence of templates and for long-range tertiary/quaternary interactions. Encouraging signs from deep evolutionary data though.

RNA modeling remains challenging for deep learning, esp. in the absence of templates and for long-range tertiary/quaternary interactions. Encouraging signs from deep evolutionary data though.
After years behind a keyboard, I will be pivoting toward the wet lab. To unlock the true potential of ML for biology/biomedicine, we need high-quality data and robust evaluation 🔬🧫🧪
After years behind a keyboard, I will be pivoting toward the wet lab. To unlock the true potential of ML for biology/biomedicine, we need high-quality data and robust evaluation 🔬🧫🧪
Paper: academic.oup.com/bioinformati...
Webserver: opig.stats.ox.ac.uk/webapps/anti...
Codebase available on Github: github.com/oxpig/AntiFold

Paper: academic.oup.com/bioinformati...
Webserver: opig.stats.ox.ac.uk/webapps/anti...
Codebase available on Github: github.com/oxpig/AntiFold

