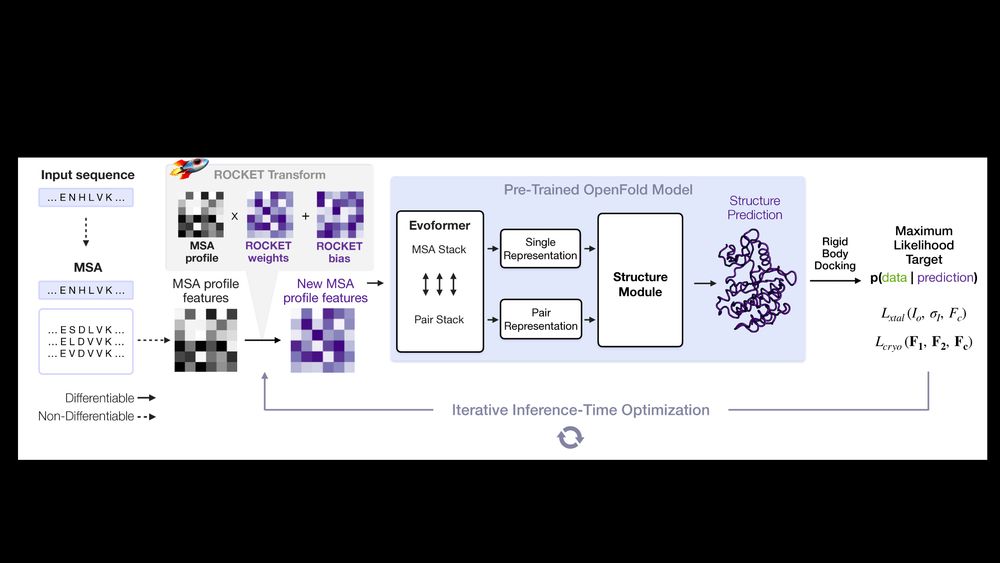
🔹 ROCKET extracts two conformations from a 9.6Å GroEL map. Its modeling matches humans here and even surpasses them in tough regions, boosting fit to data from CC=0.2 to 0.5 in a flexible domain (see ⭐) 7/14
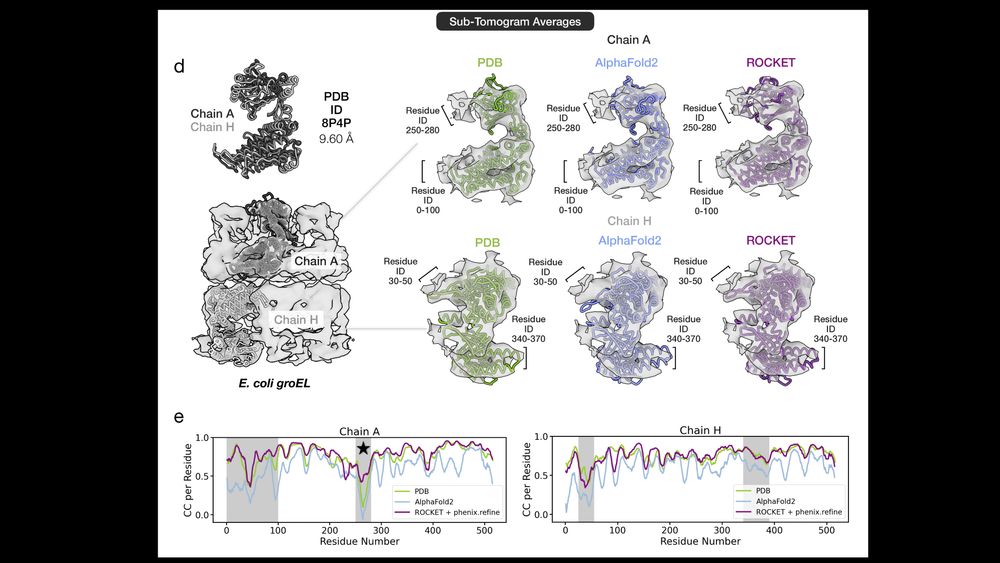
🔹 ROCKET extracts two conformations from a 9.6Å GroEL map. Its modeling matches humans here and even surpasses them in tough regions, boosting fit to data from CC=0.2 to 0.5 in a flexible domain (see ⭐) 7/14
🔹 ROCKET refines low-res (3.82 Å) HAI-1 X-ray data, improving backbone accuracy beyond AF2. It smartly preserves ambiguous regions & corrects a possibly misregistered helix (310–330), without adding geometric artifacts 6/14
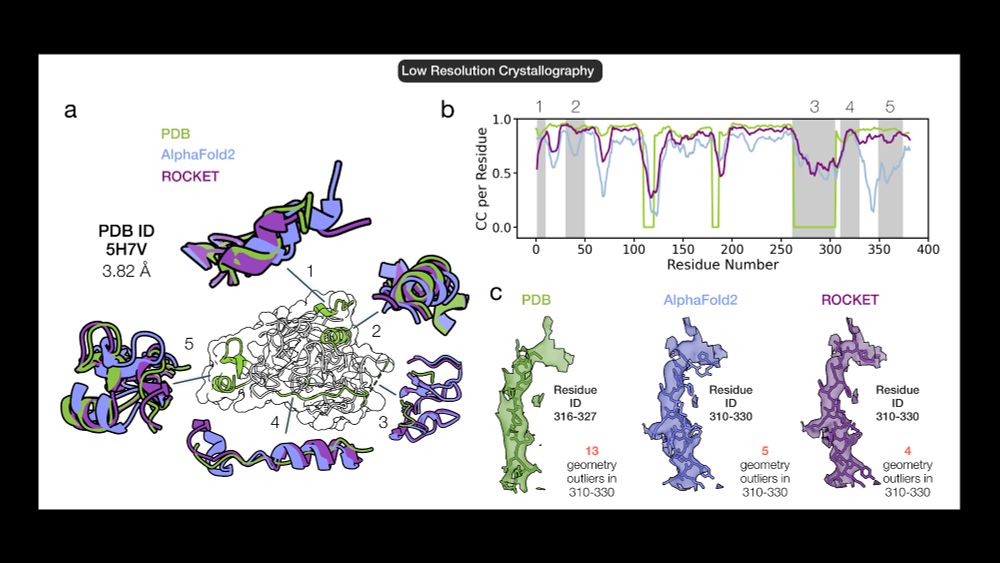
🔹 ROCKET refines low-res (3.82 Å) HAI-1 X-ray data, improving backbone accuracy beyond AF2. It smartly preserves ambiguous regions & corrects a possibly misregistered helix (310–330), without adding geometric artifacts 6/14
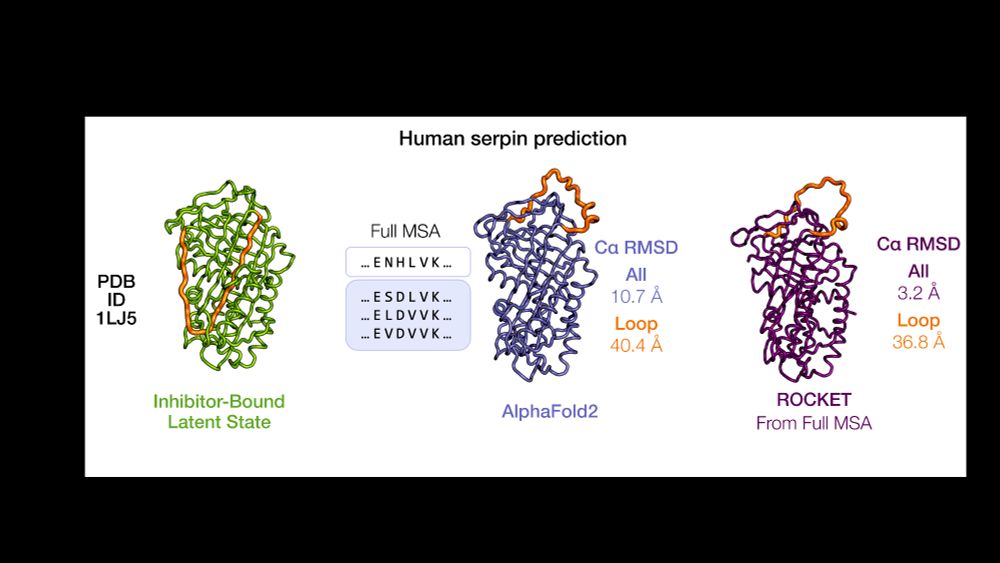
✔️ Domain shifts (e.g. GroEL)
5/14
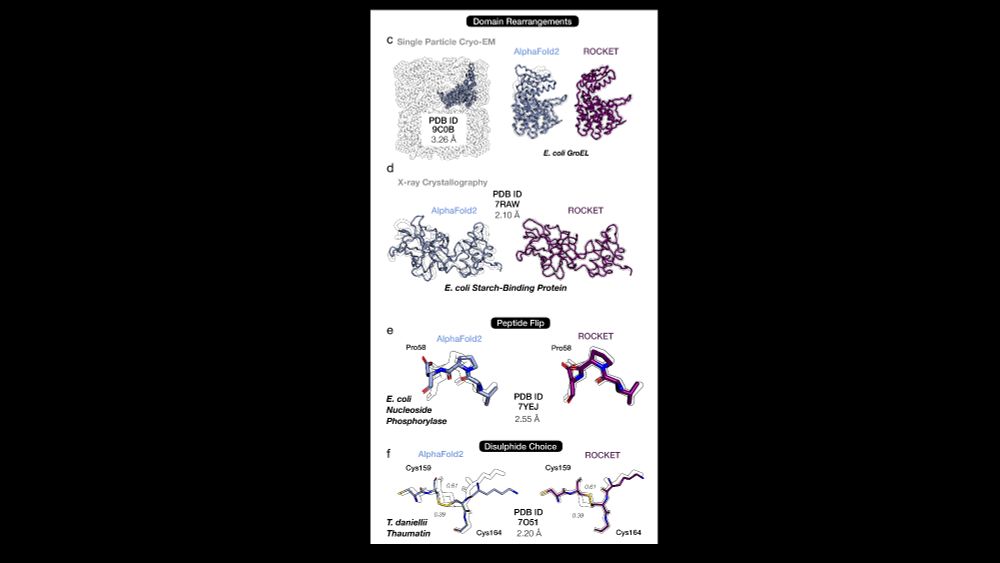
✔️ Domain shifts (e.g. GroEL)
5/14
✔️ Ligand-induced loop rearrangements (e.g. c-Abl kinase and PTP1B)
4/14
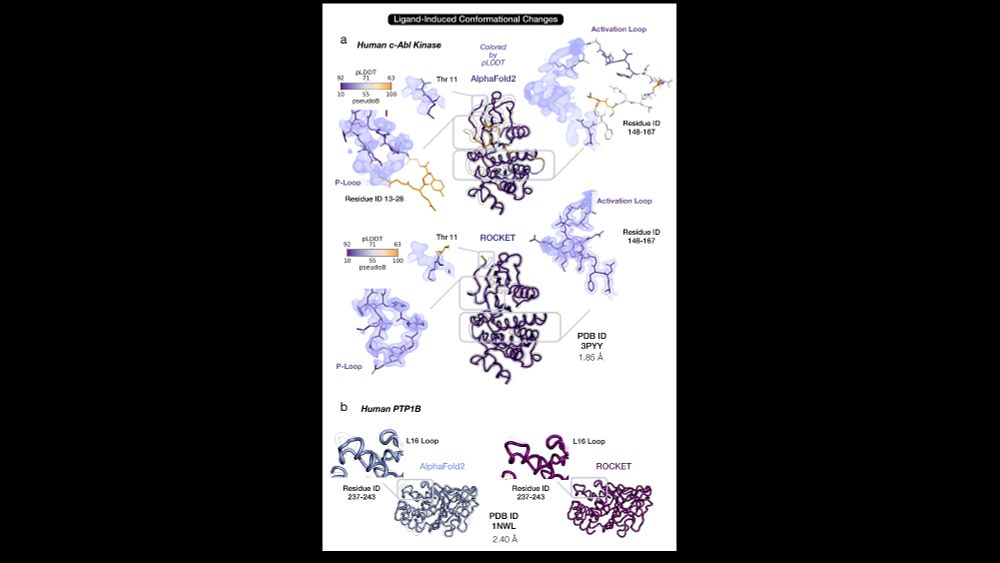
✔️ Ligand-induced loop rearrangements (e.g. c-Abl kinase and PTP1B)
4/14