Every 1.5 hours for 48 hours, we collected ocean microbes from within and adjacent to Kāneʻohe Bay, Oʻahu, Hawaiʻi and produced a multi-omics dataset.

Every 1.5 hours for 48 hours, we collected ocean microbes from within and adjacent to Kāneʻohe Bay, Oʻahu, Hawaiʻi and produced a multi-omics dataset.
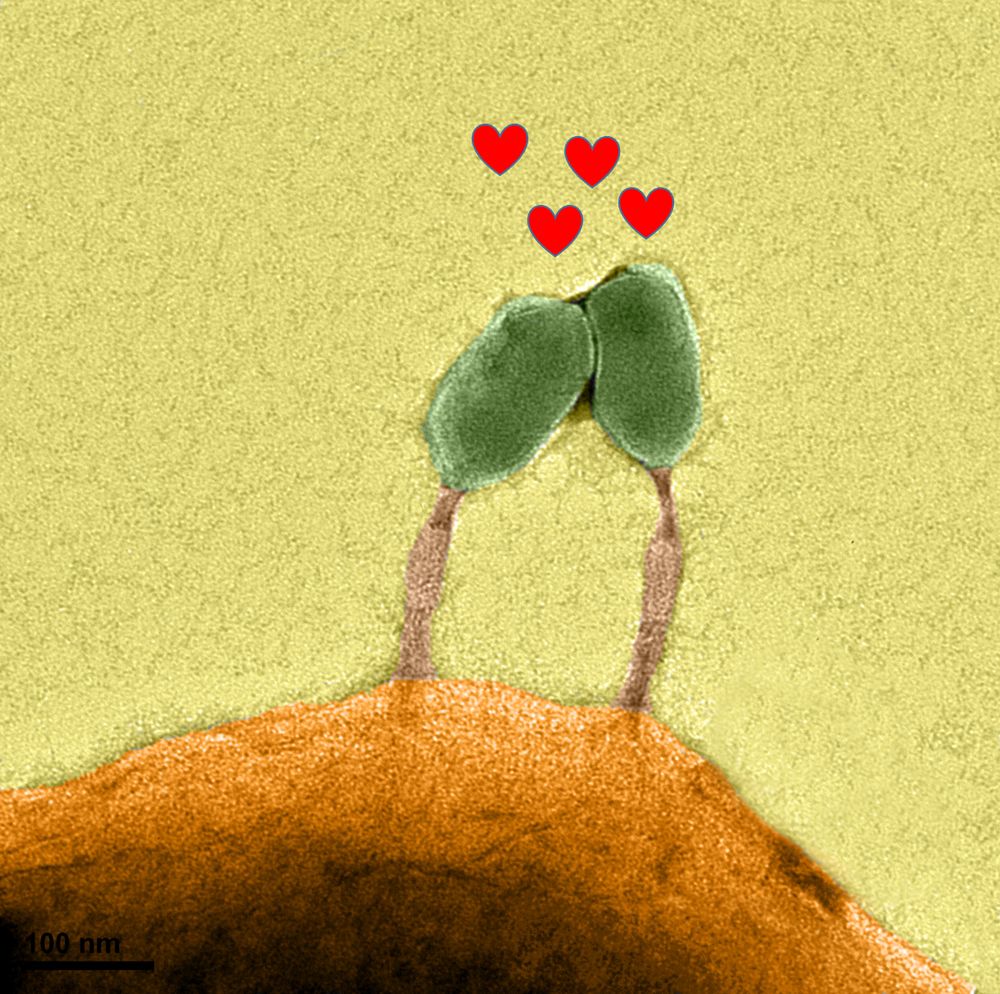
Thrilled to share our new study! We show that mirusviruses include lineages packed with spliceosomal introns and likely replicating in the nucleus of unicellular eukaryotes—a sharp contrast to most large and giant eukaryotic viruses that replicate in the cytoplasm.

Thrilled to share our new study! We show that mirusviruses include lineages packed with spliceosomal introns and likely replicating in the nucleus of unicellular eukaryotes—a sharp contrast to most large and giant eukaryotic viruses that replicate in the cytoplasm.
Please find more information on the venue, program, and the application form here, and spread the word 😇
anvio.org/workshops/20...

Please find more information on the venue, program, and the application form here, and spread the word 😇
anvio.org/workshops/20...


Here is a blog that explains my motivation for this and how to schedule a meeting:
merenlab.org/2025/11/16/E...

Here is a blog that explains my motivation for this and how to schedule a meeting:
merenlab.org/2025/11/16/E...
www.nature.com/articles/s41...

www.nature.com/articles/s41...
We present oligoN-design, a simple, reproducible and versatile open-source tool to design specific primers and probes directly from large environmental DNA datasets.
🔗 DOI: doi.org/10.1101/2025...
👉 github.com/MiguelMSandi...

We present oligoN-design, a simple, reproducible and versatile open-source tool to design specific primers and probes directly from large environmental DNA datasets.
🔗 DOI: doi.org/10.1101/2025...
👉 github.com/MiguelMSandi...
elifesciences.org/inside-elife...
I welcome that eLife is no longer indexed by SCIE. And I admire that they have not backed away from their innovative model, which makes me even more motivated to send our best work to eLife.

elifesciences.org/inside-elife...
I welcome that eLife is no longer indexed by SCIE. And I admire that they have not backed away from their innovative model, which makes me even more motivated to send our best work to eLife.


-in #ISMEJournal by Joanna Lepper, @hbrappap.bsky.social and @oliverio.bsky.social
academic.oup.com/ismej/advanc...

-in #ISMEJournal by Joanna Lepper, @hbrappap.bsky.social and @oliverio.bsky.social
academic.oup.com/ismej/advanc...
diatom induces nitrogen-fixing nodules in a
terrestrial legume
www.nature.com/articles/s41...

diatom induces nitrogen-fixing nodules in a
terrestrial legume
www.nature.com/articles/s41...

Happy to discuss applications with students whose interests align with modelling marine dispersal or oceanic forcing of coral reefs.

Happy to discuss applications with students whose interests align with modelling marine dispersal or oceanic forcing of coral reefs.
nature.com/articles/s41467-025-63463-6

nature.com/articles/s41467-025-63463-6
Ie: it's not us (Samtools team)! Be warned
The EMBL-EBI-Sanger postdoctoral fellowships might be the thing for you!
Choose one of our pre-defined topics or propose your own.
www.ebi.ac.uk/research/pos...
@sangerinstitute.bsky.social
#postdoc #sciencecareers
🧬🖥️🔬

The EMBL-EBI-Sanger postdoctoral fellowships might be the thing for you!
Choose one of our pre-defined topics or propose your own.
www.ebi.ac.uk/research/pos...
@sangerinstitute.bsky.social
#postdoc #sciencecareers
🧬🖥️🔬
Happy to share our piece "Towards a trait-based framework for protist ecology and evolution" in @cp-trendsmicrobiol.bsky.social
Let's build a unified trait 📏 database to unlock transformative insights into protist 🔬 ecology 🌍 and evolution ⏳
▶️ doi.org/10.1016/j.ti...
#protistsonsky

Happy to share our piece "Towards a trait-based framework for protist ecology and evolution" in @cp-trendsmicrobiol.bsky.social
Let's build a unified trait 📏 database to unlock transformative insights into protist 🔬 ecology 🌍 and evolution ⏳
▶️ doi.org/10.1016/j.ti...
#protistsonsky
Read more:
tmt.news/2176398

Read more:
tmt.news/2176398