doi.org/10.1021/acs....

See cgmartini.nl for details and how to apply.
Looking forward to seeing you in Groningen, Aug 11-15th.

Also check out our recent webinars on each:
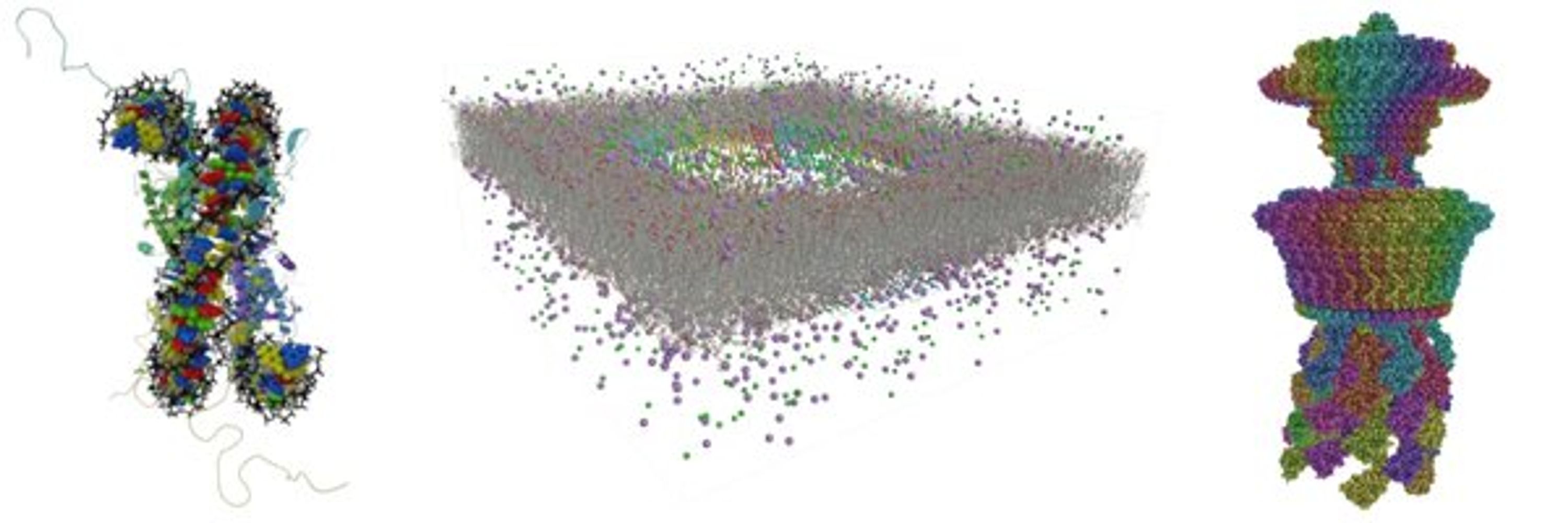
🎬 Whole cell simulation with Martini ▶️ youtu.be/fvFaPgSoM90
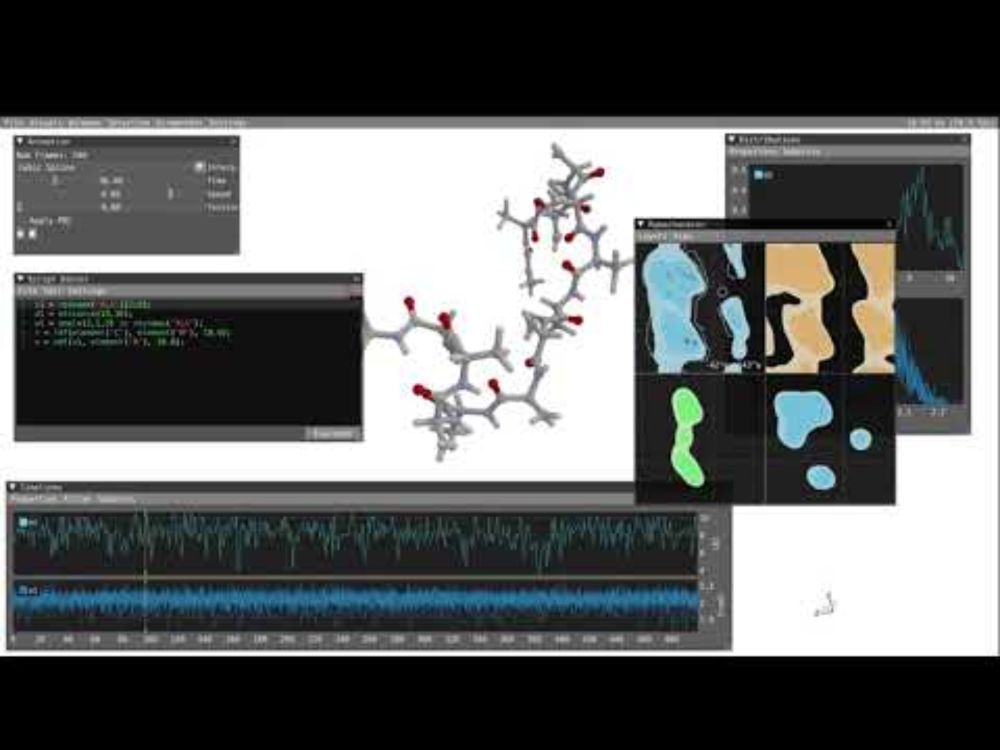
🎬 Visual analysis of #moleculardynamics with VIAMD ▶️ youtu.be/wVENzcx0XmQ
Also check out our recent webinars on each:
🎬 Whole cell simulation with Martini ▶️ youtu.be/fvFaPgSoM90
🎬 Visual analysis of #moleculardynamics with VIAMD ▶️ youtu.be/wVENzcx0XmQ

Join the VeloxChem on LUMI Workshop — 26–27 May 2025 in Espoo, Finland!
💡 Learn to:
Run VeloxChem via Jupyter
Scale jobs on LUMI's GPUs with Slurm
Analyze results with VIAMD
Info + signup 👉 lumi-supercomputer.eu/events/velox...
lumi-supercomputer.eu/events/velox...

Join the VeloxChem on LUMI Workshop — 26–27 May 2025 in Espoo, Finland!
💡 Learn to:
Run VeloxChem via Jupyter
Scale jobs on LUMI's GPUs with Slurm
Analyze results with VIAMD
Info + signup 👉 lumi-supercomputer.eu/events/velox...
lumi-supercomputer.eu/events/velox...



