
Rafael Peña-Miller
@penamiller.bsky.social
Scientist working somewhere around mathematical, computational and experimental microbiology
Reposted by Rafael Peña-Miller
New paper with my (amazing) friend and mentor @jrpenades.bsky.social
Really looking forward to see what plasmid aficionados think of this one!!
With @asantoslopez.bsky.social @wfigueroac3.bsky.social Akshay Sabins and others
www.cell.com/cell-reports...
Really looking forward to see what plasmid aficionados think of this one!!
With @asantoslopez.bsky.social @wfigueroac3.bsky.social Akshay Sabins and others
www.cell.com/cell-reports...

Non-conjugative plasmids limit their mobility to persist in nature
Sabnis et al. explain why non-conjugative plasmids move at a low rate in nature. While
increased mobility can easily evolve by incorporating phage DNA into plasmids, this
is disadvantageous because it...
www.cell.com
October 22, 2025 at 1:12 PM
New paper with my (amazing) friend and mentor @jrpenades.bsky.social
Really looking forward to see what plasmid aficionados think of this one!!
With @asantoslopez.bsky.social @wfigueroac3.bsky.social Akshay Sabins and others
www.cell.com/cell-reports...
Really looking forward to see what plasmid aficionados think of this one!!
With @asantoslopez.bsky.social @wfigueroac3.bsky.social Akshay Sabins and others
www.cell.com/cell-reports...
Reposted by Rafael Peña-Miller
Do plasmids evolve faster 🐇, slower 🐢, or just like chromosomes 🧬?
In our new paper, we tackled this question using theory, simulations, bioinformatics, and experiments!
👇 Check out all the details in Paula’s thread!
Hint: 🐇 (most of the time)
In our new paper, we tackled this question using theory, simulations, bioinformatics, and experiments!
👇 Check out all the details in Paula’s thread!
Hint: 🐇 (most of the time)
July 22, 2025 at 3:05 PM
Do plasmids evolve faster 🐇, slower 🐢, or just like chromosomes 🧬?
In our new paper, we tackled this question using theory, simulations, bioinformatics, and experiments!
👇 Check out all the details in Paula’s thread!
Hint: 🐇 (most of the time)
In our new paper, we tackled this question using theory, simulations, bioinformatics, and experiments!
👇 Check out all the details in Paula’s thread!
Hint: 🐇 (most of the time)
Reposted by Rafael Peña-Miller
New paper alert! 🚨
Plasmids promote bacterial evolution through a copy number-driven increase in mutation rate.
We combine theory, simulations, experimental evolution, and bioinformatics to demonstrate that mutation rates scale with plasmid copy number.
Let's dive in! 🧵👇
Plasmids promote bacterial evolution through a copy number-driven increase in mutation rate.
We combine theory, simulations, experimental evolution, and bioinformatics to demonstrate that mutation rates scale with plasmid copy number.
Let's dive in! 🧵👇

Plasmids promote bacterial evolution through a copy number-driven increase in mutation rate
Plasmids are autonomously replicating DNA molecules that stably coexist with chromosomes in bacterial cells. These genetic elements drive horizontal gene transfer and play a fundamental role in bacter...
www.biorxiv.org
July 22, 2025 at 9:38 AM
New paper alert! 🚨
Plasmids promote bacterial evolution through a copy number-driven increase in mutation rate.
We combine theory, simulations, experimental evolution, and bioinformatics to demonstrate that mutation rates scale with plasmid copy number.
Let's dive in! 🧵👇
Plasmids promote bacterial evolution through a copy number-driven increase in mutation rate.
We combine theory, simulations, experimental evolution, and bioinformatics to demonstrate that mutation rates scale with plasmid copy number.
Let's dive in! 🧵👇
Reposted by Rafael Peña-Miller
🚨🚨New paper out in @natcomms.nature.com!!
Come for the first large-scale analysis of plasmid copy number across species,
stay for one of the most intriguing results of my lab: universal scaling laws in plasmid biology! 📈🧬
👉 www.nature.com/articles/s41...
Come for the first large-scale analysis of plasmid copy number across species,
stay for one of the most intriguing results of my lab: universal scaling laws in plasmid biology! 📈🧬
👉 www.nature.com/articles/s41...
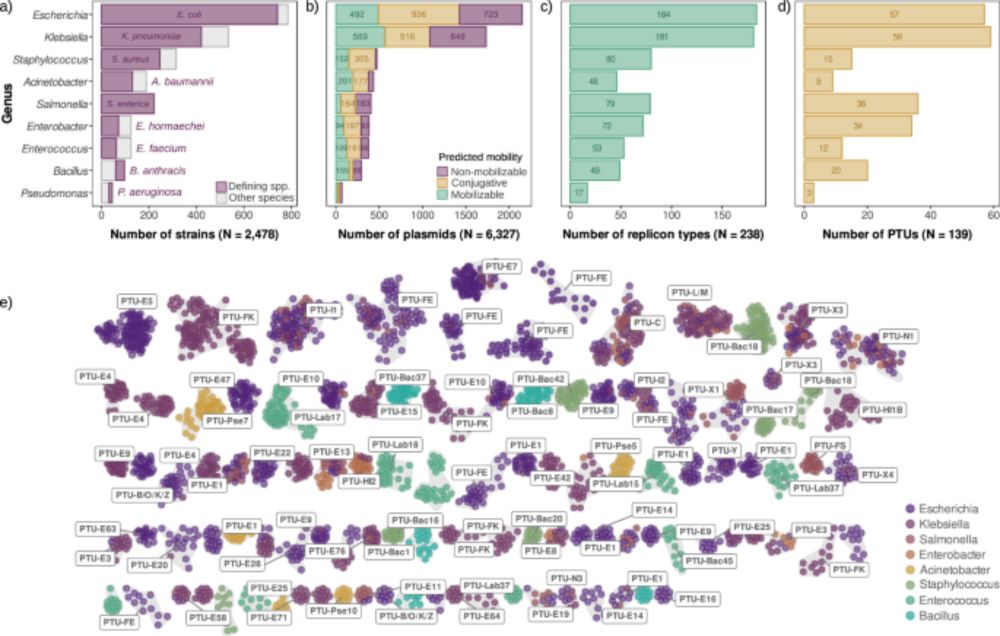
Universal rules govern plasmid copy number - Nature Communications
Plasmids exhibit a broad range of sizes and copies per cell, and these two parameters appear to be negatively correlated. Here, Ramiro-Martínez et al. analyse the copy number of thousands of diverse b...
www.nature.com
July 2, 2025 at 11:08 AM
🚨🚨New paper out in @natcomms.nature.com!!
Come for the first large-scale analysis of plasmid copy number across species,
stay for one of the most intriguing results of my lab: universal scaling laws in plasmid biology! 📈🧬
👉 www.nature.com/articles/s41...
Come for the first large-scale analysis of plasmid copy number across species,
stay for one of the most intriguing results of my lab: universal scaling laws in plasmid biology! 📈🧬
👉 www.nature.com/articles/s41...
New preprint led by @brunoluviano.bsky.social & Fernando Santos.
We show that filamentation enhances bacterial survival under toxic stress — not as collateral damage, but as a regulated morphological response.
TL;DR: Filamentation isn’t a symptom, it’s a strategy!
www.biorxiv.org/content/10.1...
We show that filamentation enhances bacterial survival under toxic stress — not as collateral damage, but as a regulated morphological response.
TL;DR: Filamentation isn’t a symptom, it’s a strategy!
www.biorxiv.org/content/10.1...
May 15, 2025 at 5:23 PM
New preprint led by @brunoluviano.bsky.social & Fernando Santos.
We show that filamentation enhances bacterial survival under toxic stress — not as collateral damage, but as a regulated morphological response.
TL;DR: Filamentation isn’t a symptom, it’s a strategy!
www.biorxiv.org/content/10.1...
We show that filamentation enhances bacterial survival under toxic stress — not as collateral damage, but as a regulated morphological response.
TL;DR: Filamentation isn’t a symptom, it’s a strategy!
www.biorxiv.org/content/10.1...
Reposted by Rafael Peña-Miller
🧪
Finally out after peer review, our work showing that "Mobile #Integrons carry Phage Defense Systems" is now published in Science 🎉
Short 🧵
www.science.org/doi/10.1126/...
Finally out after peer review, our work showing that "Mobile #Integrons carry Phage Defense Systems" is now published in Science 🎉
Short 🧵
www.science.org/doi/10.1126/...

Mobile integrons encode phage defense systems
Integrons are bacterial genetic elements that capture, stockpile, and modulate the expression of genes encoded in integron cassettes. Mobile integrons (MIs) are borne on plasmids, acting as a vehicle ...
www.science.org
May 8, 2025 at 8:27 PM
🧪
Finally out after peer review, our work showing that "Mobile #Integrons carry Phage Defense Systems" is now published in Science 🎉
Short 🧵
www.science.org/doi/10.1126/...
Finally out after peer review, our work showing that "Mobile #Integrons carry Phage Defense Systems" is now published in Science 🎉
Short 🧵
www.science.org/doi/10.1126/...
Reposted by Rafael Peña-Miller
We are recruiting! We are looking for a PhD student to study rapid adaptation to stress in bacteria (shorturl.at/sr0Mj). Come join us in Barcelona!
📣 Exciting News! The Evolutionary Microbiology Lab is hiring:
📍 The #IBE seeks a PhD Student.
📆 Application Deadline: 31st of March.
#JoinTheIBE and be part of our team!
✍️https://www.ibe.upf-csic.es/work-with-us/job-offers
📍 The #IBE seeks a PhD Student.
📆 Application Deadline: 31st of March.
#JoinTheIBE and be part of our team!
✍️https://www.ibe.upf-csic.es/work-with-us/job-offers

February 21, 2025 at 2:03 PM
We are recruiting! We are looking for a PhD student to study rapid adaptation to stress in bacteria (shorturl.at/sr0Mj). Come join us in Barcelona!
Reposted by Rafael Peña-Miller
Check out this amazing Spotlight on our recently published paper -> www.cell.com/trends/micro...
Huge thanks to @oliviakosterlitz.bsky.social, Benjamin Kerr and Elisabeth Duan for this great spolight.
Huge thanks to @oliviakosterlitz.bsky.social, Benjamin Kerr and Elisabeth Duan for this great spolight.
November 20, 2024 at 4:08 PM
Check out this amazing Spotlight on our recently published paper -> www.cell.com/trends/micro...
Huge thanks to @oliviakosterlitz.bsky.social, Benjamin Kerr and Elisabeth Duan for this great spolight.
Huge thanks to @oliviakosterlitz.bsky.social, Benjamin Kerr and Elisabeth Duan for this great spolight.
Our paper on plasmid-driven heterogeneity is finally out!
www.nature.com/articles/s41...
@sanmillan.bsky.social @ayari.bsky.social @jerorb.bsky.social @craigmaclean.bsky.social
www.nature.com/articles/s41...
@sanmillan.bsky.social @ayari.bsky.social @jerorb.bsky.social @craigmaclean.bsky.social
March 25, 2024 at 5:26 PM
Our paper on plasmid-driven heterogeneity is finally out!
www.nature.com/articles/s41...
@sanmillan.bsky.social @ayari.bsky.social @jerorb.bsky.social @craigmaclean.bsky.social
www.nature.com/articles/s41...
@sanmillan.bsky.social @ayari.bsky.social @jerorb.bsky.social @craigmaclean.bsky.social
Reposted by Rafael Peña-Miller
Thanks Thanks to @ayari.bsky.social for inviting me to present at @meehubs.bsky.social from the Cuernavaca hub. Got some great questions from the audience in other parts of the World!
January 10, 2024 at 8:46 PM
Thanks Thanks to @ayari.bsky.social for inviting me to present at @meehubs.bsky.social from the Cuernavaca hub. Got some great questions from the audience in other parts of the World!
Reposted by Rafael Peña-Miller
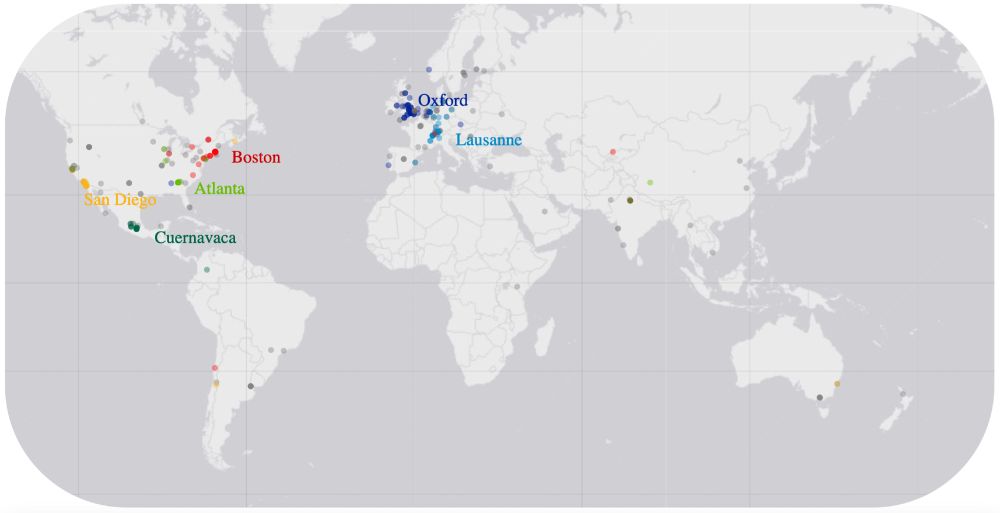
Only a bit more than 12 hours till start of #MEEhubs2024. We have hundreds of registered participants, some shared their location and where they travel to on the map below! #MicroSky
January 8, 2024 at 7:03 PM
Only a bit more than 12 hours till start of #MEEhubs2024. We have hundreds of registered participants, some shared their location and where they travel to on the map below! #MicroSky
Reposted by Rafael Peña-Miller
Can we predict AMR evolution?? We sure can try! New study published today in PNAS
www.pnas.org/doi/10.1073/...
www.pnas.org/doi/10.1073/...
December 14, 2023 at 9:09 PM
Can we predict AMR evolution?? We sure can try! New study published today in PNAS
www.pnas.org/doi/10.1073/...
www.pnas.org/doi/10.1073/...

