Huge collaborative effort from many people
Check out the whole story here
Huge collaborative effort from many people
Check out the whole story here
Detecting Protein Higher-Order Structural Changes Using Kinase as a Phospho-Labeler www.biorxiv.org/content/10.1...

Detecting Protein Higher-Order Structural Changes Using Kinase as a Phospho-Labeler www.biorxiv.org/content/10.1...
www.nature.com/articles/s41...
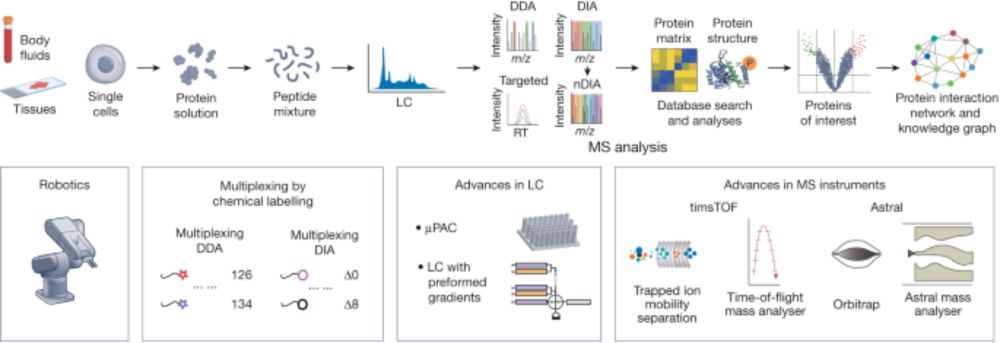
www.nature.com/articles/s41...

go.bsky.app/HH7kqEh
go.bsky.app/HH7kqEh
Feel free to ask to be added or removed, I've tried to make a guess but could've got it wrong.
I think it's good to also have an ECR network.
#teamMassSpec
go.bsky.app/Dp8PHX1
Feel free to ask to be added or removed, I've tried to make a guess but could've got it wrong.
I think it's good to also have an ECR network.
#teamMassSpec
go.bsky.app/Dp8PHX1

