Currently in the Bock lab@CeMM talking to single cells with LLMs.
Here: paper reviews in 3 lines
Mastodon (inactive): @moritzschaefer@qoto.org
Twitter: @muronglizi
Try it yourself: cellwhisperer.bocklab.org and check out our tweetorial.
Huge shout-out to to all the other contributors for the amazing teamwork!
Try it yourself: cellwhisperer.bocklab.org and check out our tweetorial.
Huge shout-out to to all the other contributors for the amazing teamwork!
SpotWhisperer enables spatially resolved annotation of histopathology images using natural language. We achieved this by "transferring" annotations from transcriptomic data. More soon!

SpotWhisperer enables spatially resolved annotation of histopathology images using natural language. We achieved this by "transferring" annotations from transcriptomic data. More soon!
I am grateful for the friendships I made and the science I got to contribute to.
I also wrote a brief recap of my science and activities during that time. Enjoy! moritzs.de/cemm
I am grateful for the friendships I made and the science I got to contribute to.
I also wrote a brief recap of my science and activities during that time. Enjoy! moritzs.de/cemm
Made possible by an amazing bunch of colleagues at my institute.
Made possible by an amazing bunch of colleagues at my institute.
hrovatin.github.io/posts/sc_flo...

hrovatin.github.io/posts/sc_flo...
>500 people in front of @univie.ac.at, ~1000 people at Votivpark! 💚💚💚
Science is for everyone!
#standupforscience2025
#scienceforall
#sciencenotsilence

>500 people in front of @univie.ac.at, ~1000 people at Votivpark! 💚💚💚
Science is for everyone!
#standupforscience2025
#scienceforall
#sciencenotsilence
Here's our list of current speakers—and it's still growing! Come tomorrow to hear some amazing speakers talk about science, their experiences, and the future.
#standupforscience2025

Here's our list of current speakers—and it's still growing! Come tomorrow to hear some amazing speakers talk about science, their experiences, and the future.
#standupforscience2025
LLMs should rather be seen as "interface modules" of more complex AI systems: Their core role will be to *translate* between AI language (bits and bytes) and human-readable language.
LLMs should rather be seen as "interface modules" of more complex AI systems: Their core role will be to *translate* between AI language (bits and bytes) and human-readable language.
- My highlights post on LinkedIn:
www.linkedin.com/posts/moritz...
- I gave a talk too: lnkd.in/dXRF2mai
- All talks here: media.ccc.de
- My highlights post on LinkedIn:
www.linkedin.com/posts/moritz...
- I gave a talk too: lnkd.in/dXRF2mai
- All talks here: media.ccc.de
1. Research associate (wet-lab w/ phd) to generate mpra perturbation data
2. ML postdoc to build multimodal generative AI for DNA (eg diffusion and LLMs)
3. Bioinformatician (any level) to process and harmonize functional genomics data to train foundation models
DM me if interested!
1. Research associate (wet-lab w/ phd) to generate mpra perturbation data
2. ML postdoc to build multimodal generative AI for DNA (eg diffusion and LLMs)
3. Bioinformatician (any level) to process and harmonize functional genomics data to train foundation models
DM me if interested!
www.newsweek.com/elon-musk-ta...

www.newsweek.com/elon-musk-ta...
It's a really great read and builds on an interview I had with the @kamalnahas.bsky.social.
www.the-scientist.com/an-ai-lab-pa...

It's a really great read and builds on an interview I had with the @kamalnahas.bsky.social.
www.the-scientist.com/an-ai-lab-pa...
Thanks @mike_schatz and all organizers, looking forward to being back in 2026!

Thanks @mike_schatz and all organizers, looking forward to being back in 2026!
I thought it dead already.
I thought it dead already.
#ProudPI #stats #spatial #omics #transferlearning 🧪🧬🖥️🧠📈

#ProudPI #stats #spatial #omics #transferlearning 🧪🧬🖥️🧠📈
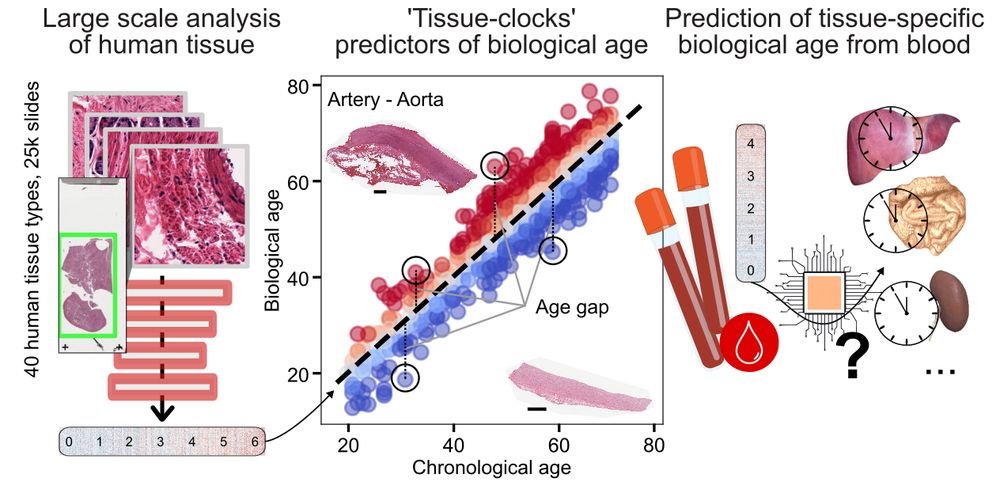
- Protein embeddings are powerful.
- But: they are usually cell-type agnostic
- This paper: Embedding cell-type-aware protein representations via GNNs on scRNA-seq-pruned protein interaction graphs
www.nature.com/articles/s41...

- Protein embeddings are powerful.
- But: they are usually cell-type agnostic
- This paper: Embedding cell-type-aware protein representations via GNNs on scRNA-seq-pruned protein interaction graphs
www.nature.com/articles/s41...
Information over-squashing in language tasks"
- In typical LLMs every token only 'sees' previous tokens (causal attention)
- This leads to information inbalance across tokens
- Thereby, later tokens 'vanish' in long sequences
arxiv.org/pdf/2406.04267
Information over-squashing in language tasks"
- In typical LLMs every token only 'sees' previous tokens (causal attention)
- This leads to information inbalance across tokens
- Thereby, later tokens 'vanish' in long sequences
arxiv.org/pdf/2406.04267
- PrE is extra-embryonic tissue (at day 4) but can still recreate a full blastocyst (i.e. also embryo)
- JAK/STAT supports this plasticity
- www.cell.com/cell/abstrac...
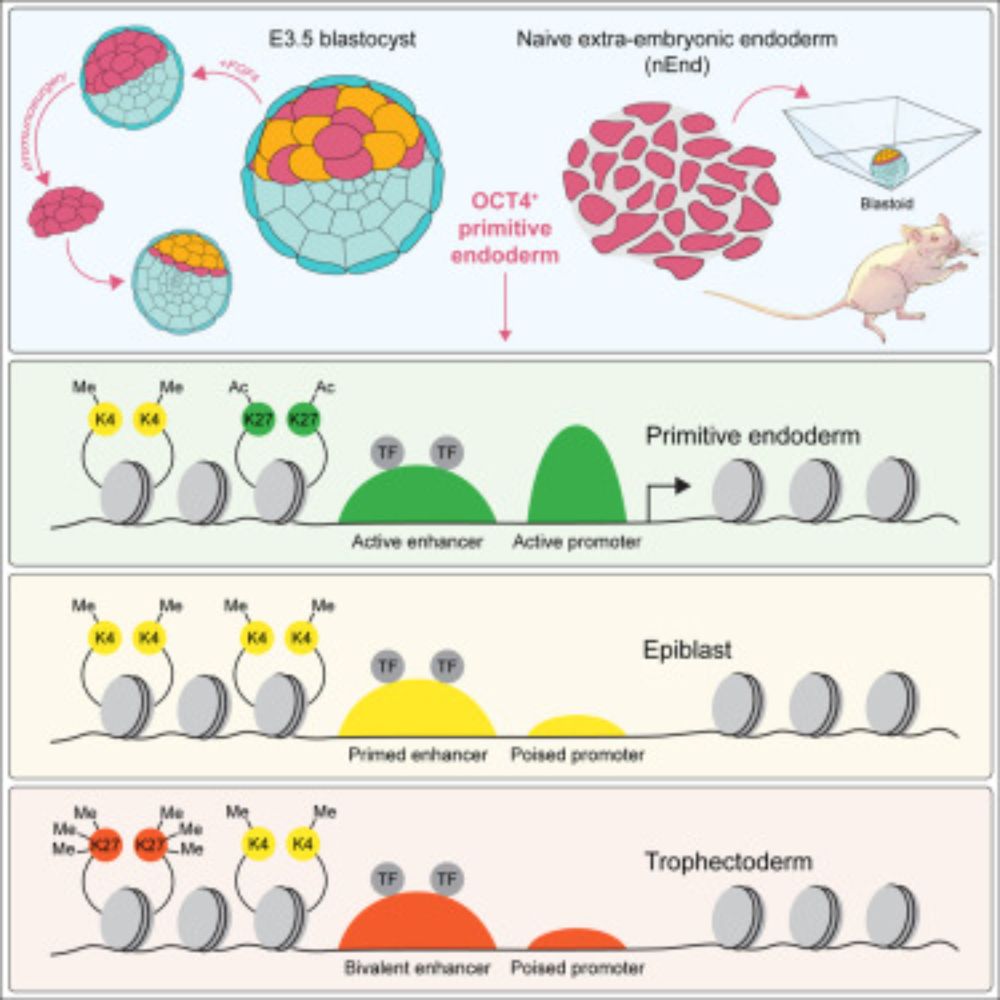
- PrE is extra-embryonic tissue (at day 4) but can still recreate a full blastocyst (i.e. also embryo)
- JAK/STAT supports this plasticity
- www.cell.com/cell/abstrac...
- Humans, as hunter-gatherers, evolved to 'always move'
- Exercise alters gene expression throughout the body
- With exercise, muscles contribute to anti-inflammation through IL-6 production
www.nature.com/articles/d41... 🧬🖥️🧪

- Humans, as hunter-gatherers, evolved to 'always move'
- Exercise alters gene expression throughout the body
- With exercise, muscles contribute to anti-inflammation through IL-6 production
www.nature.com/articles/d41... 🧬🖥️🧪
Hats off to my UCR colleagues Kevin Esterling and Diogo Ferrari and their co-authors...
🧪sociology polisky policysky
www.nature.com/articles/s41...

Hats off to my UCR colleagues Kevin Esterling and Diogo Ferrari and their co-authors...
🧪sociology polisky policysky
www.nature.com/articles/s41...
- Most drug design methods assume rigid proteins -
Simple docking experiments show: Flexible side chains enhance modeling
- New method FlexFlow: Flow matching of small molecules in addition to side chain configurations
Looking forward to full release 🧬 & 🖥️
- Most drug design methods assume rigid proteins -
Simple docking experiments show: Flexible side chains enhance modeling
- New method FlexFlow: Flow matching of small molecules in addition to side chain configurations
Looking forward to full release 🧬 & 🖥️
One more example of how polarisation wins the attention market..
One more example of how polarisation wins the attention market..
- Here, GPT- and protein embeddings enable perturbation effect prediction for unseen genes
- My take: Great direction and I anticipate more 'embedding arithmetics' (e.g. 'gene1-gene2' for overexpression and KO)

- Here, GPT- and protein embeddings enable perturbation effect prediction for unseen genes
- My take: Great direction and I anticipate more 'embedding arithmetics' (e.g. 'gene1-gene2' for overexpression and KO)
- In GNNs, when data needs to pass many nodes, the data gets exponentially 'diluted'
- Adding a simply fully-connected edge layer, improved upon QM9 by 42%
- This foundational and simple concept is only from 3 years ago

- In GNNs, when data needs to pass many nodes, the data gets exponentially 'diluted'
- Adding a simply fully-connected edge layer, improved upon QM9 by 42%
- This foundational and simple concept is only from 3 years ago

