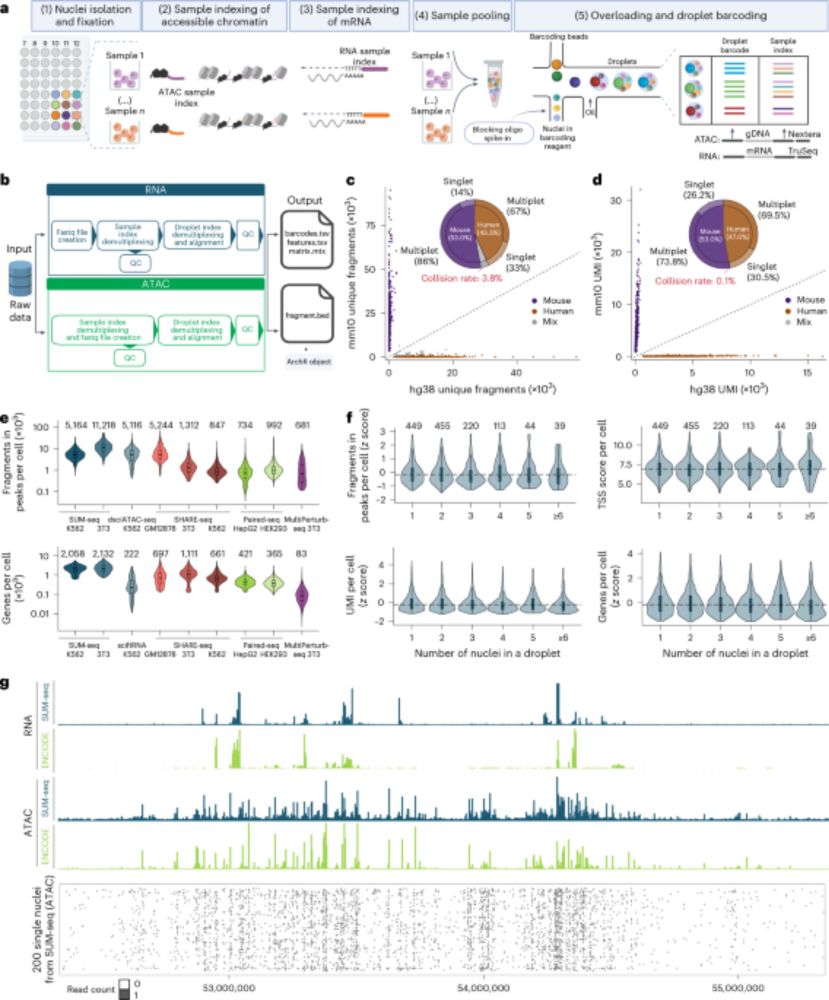
🚀 Ultra-high-throughput Multiplexed snATAC+RNA
Used to:
⏳ link temporal macrophage GRNs to immune disease genetics
🩸 map T cell regulatory landscapes
🧬✂️ dissect TF function in hiPSC differentiation via CRISPRi/a screens
doi.org/10.1038/s41592-025-02700-8
🧵
Happy to been part of the single cell tech dev/analysis of this study lead by @umut_yildiz12 from EMBL/Noh lab! www.biorxiv.org/content/10.1...
Happy to been part of the single cell tech dev/analysis of this study lead by @umut_yildiz12 from EMBL/Noh lab! www.biorxiv.org/content/10.1...
www.nature.com/articles/s41...

www.nature.com/articles/s41...
Congrats Prof. Zaugg & collaborators!
🔗 www.nature.com/articles/s41...

Congrats Prof. Zaugg & collaborators!
🔗 www.nature.com/articles/s41...
🚀 Ultra-high-throughput Multiplexed snATAC+RNA
Used to:
⏳ link temporal macrophage GRNs to immune disease genetics
🩸 map T cell regulatory landscapes
🧬✂️ dissect TF function in hiPSC differentiation via CRISPRi/a screens
doi.org/10.1038/s41592-025-02700-8
🧵

🚀 Ultra-high-throughput Multiplexed snATAC+RNA
Used to:
⏳ link temporal macrophage GRNs to immune disease genetics
🩸 map T cell regulatory landscapes
🧬✂️ dissect TF function in hiPSC differentiation via CRISPRi/a screens
doi.org/10.1038/s41592-025-02700-8
🧵
biorxiv.org/content/10.1101/2024.12.11.627935v1
biorxiv.org/content/10.1101/2024.12.11.627935v1
Perfect if you want to scale up time course, drug/CRISPR screen or atlas projects! 🖥️ 🧬
www.biorxiv.org/content/10.1...
Perfect if you want to scale up time course, drug/CRISPR screen or atlas projects! 🖥️ 🧬
www.biorxiv.org/content/10.1...

