University of Rochester. RNA-targeting CRISPR / CRISPR membrane proteins / RNA acetylation. An Aussie in 'the land of the free'. Music Nerd
@doc-jlmeier.bsky.social , Colin Wu and Schraga Schwartz labs among others, where we try to figure out the role of ac4C tRNA mods in mammalian stress signaling. See below for an excellent summary from Jordan (thanks!)
www.science.org/doi/10.1126/...
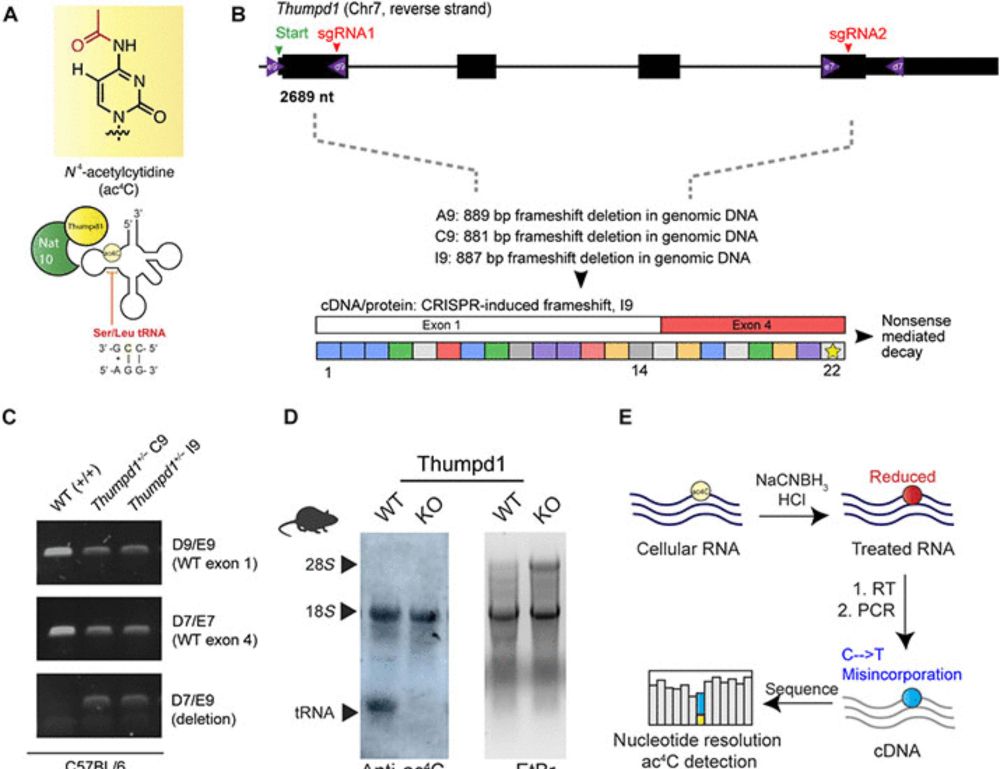
@doc-jlmeier.bsky.social , Colin Wu and Schraga Schwartz labs among others, where we try to figure out the role of ac4C tRNA mods in mammalian stress signaling. See below for an excellent summary from Jordan (thanks!)


