computational biology, ML, protein design, cheminformatics, fancy dev tooling, tinge of bouldering
https://marinegor.dev
1. Search literature (currently stubbed)
2. Enumerate papers, extract contacts
3. Send email w/ data drop location
4. Parse data
Does anyone want to help productionize this?
1. Search literature (currently stubbed)
2. Enumerate papers, extract contacts
3. Send email w/ data drop location
4. Parse data
Does anyone want to help productionize this?


🗓️ November 9-11, 2025
📍Tempe, Arizona, USA (and online)
#open-source-software #molecular #simulations #streaming

🗓️ November 9-11, 2025
📍Tempe, Arizona, USA (and online)
#open-source-software #molecular #simulations #streaming
For uv users, just do:
```
uv run gist.githubusercontent.com/marinegor/86...
```
For uv users, just do:
```
uv run gist.githubusercontent.com/marinegor/86...
```
Includes:
* Additional Gromacs/distopia/parallel analysis support
* New "water"/"precision" keywords
🙏 to the release's 10 contributors (3 new), and to @numfocus.bsky.social/@chanzuckerberg.bsky.social for support.
Includes:
* Additional Gromacs/distopia/parallel analysis support
* New "water"/"precision" keywords
🙏 to the release's 10 contributors (3 new), and to @numfocus.bsky.social/@chanzuckerberg.bsky.social for support.
Read our blog on how to apply: www.mdanalysis.org/2025/02/28/g.... Pre-proposals are due March 21st.
We value transparency; please communicate in public forums.

Read our blog on how to apply: www.mdanalysis.org/2025/02/28/g.... Pre-proposals are due March 21st.
We value transparency; please communicate in public forums.

Also covers variants like non-Euclidean & discrete flow matching.
A PyTorch library is also released with this guide!
This looks like a very good read! 🔥
arxiv: arxiv.org/abs/2412.06264

Also covers variants like non-Euclidean & discrete flow matching.
A PyTorch library is also released with this guide!
This looks like a very good read! 🔥
arxiv: arxiv.org/abs/2412.06264
I remember how the paper about Zipf law for small molecule's substructures actually blew my mind, this gets pretty close too :)
arxiv.org/abs/2411.17669
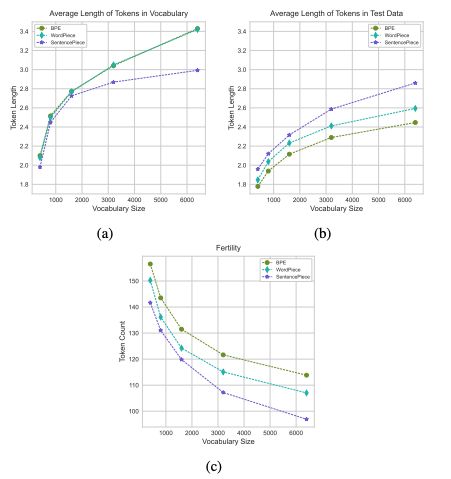
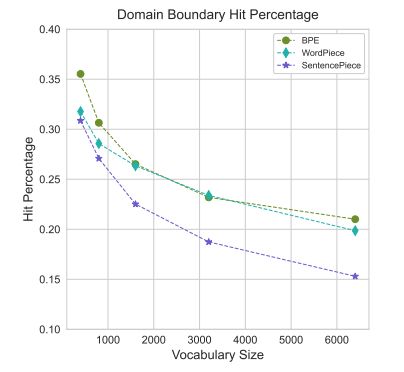
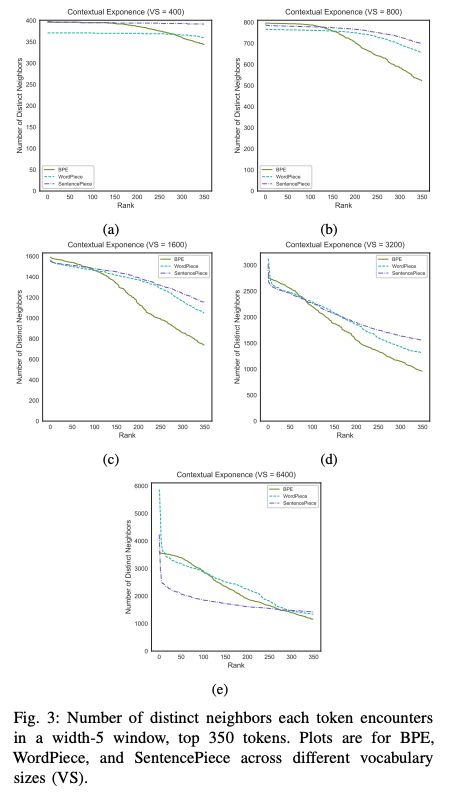
I remember how the paper about Zipf law for small molecule's substructures actually blew my mind, this gets pretty close too :)

github.com/MDAnalysis/m...
It's been there for a while now but isn't still in a tagged version afaik, so you have to check it out manually to use.

github.com/MDAnalysis/m...
It's been there for a while now but isn't still in a tagged version afaik, so you have to check it out manually to use.
Anyway, hi everyone, I'm happy to (re)connect with everyone I know and don't know -- I'll post some old-but-gold things about myself soon, stay tuned!
Anyway, hi everyone, I'm happy to (re)connect with everyone I know and don't know -- I'll post some old-but-gold things about myself soon, stay tuned!
Spots are limited, so make sure to apply before Feb 19: www.mdanalysis.org/2024/02/05/m...

Spots are limited, so make sure to apply before Feb 19: www.mdanalysis.org/2024/02/05/m...
...and hi bsky, I guess?🤔
www.mdanalysis.org/2024/01/18/g...
Also, please get in touch with us if you are interested in #mentoring with MDAnalysis for the GSoC 2024 program!

...and hi bsky, I guess?🤔

