ML🤖+Bio🧬 https://brbiclab.epfl.ch/
Systema helps to evaluate perturbation response prediction methods by focusing on perturbation-specific effects rather than systematic variation 🎯

Systema helps to evaluate perturbation response prediction methods by focusing on perturbation-specific effects rather than systematic variation 🎯
Preprint 📝: biorxiv.org/content/10.1...
🧵 1/n
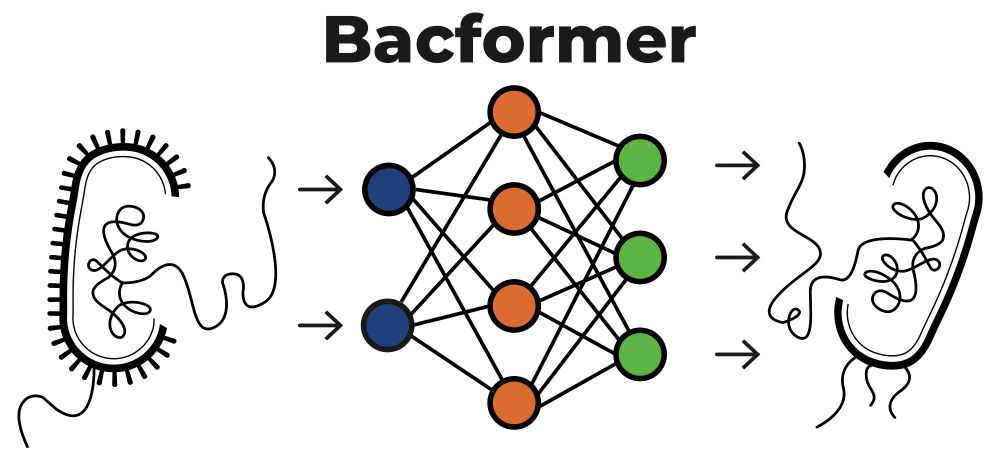
Preprint 📝: biorxiv.org/content/10.1...
🧵 1/n
Join us for the DREAM Challenges session on Monday, 21 July, 14:00 Explore community-driven benchmarking for Foundation Models—run a live experiment for LLMs.
@bowang87.bsky.social @anshulkundaje.bsky.social @cziscience.bsky.social @mariabrbic.bsky.social @juliosaezrod.bsky.social

Join us for the DREAM Challenges session on Monday, 21 July, 14:00 Explore community-driven benchmarking for Foundation Models—run a live experiment for LLMs.
@bowang87.bsky.social @anshulkundaje.bsky.social @cziscience.bsky.social @mariabrbic.bsky.social @juliosaezrod.bsky.social
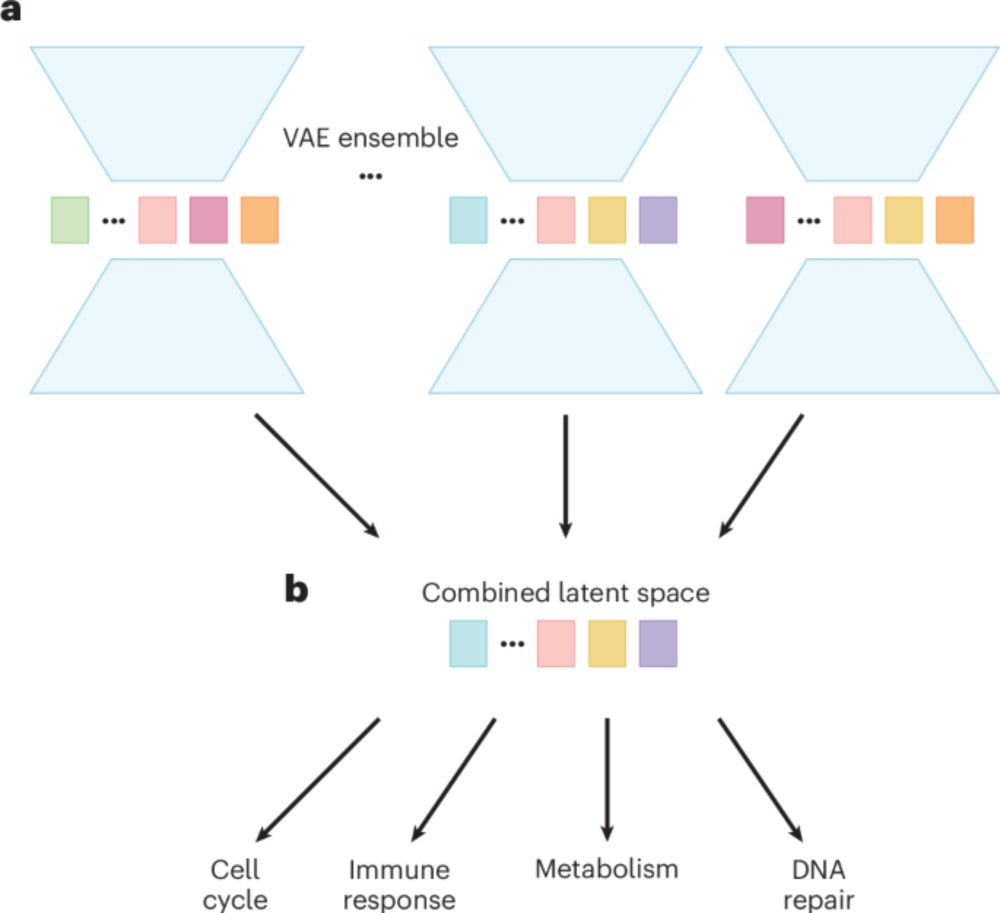
Check out our paper here: www.biorxiv.org/content/10.1...
Check out our paper here: www.biorxiv.org/content/10.1...
@weiqiu.bsky.social is on the academic job market!
@weiqiu.bsky.social is on the academic job market!
www.encodeproject.org/single-cell/...
www.encodeproject.org/single-cell/...
Paper: doi.org/10.1101/2024.12.20.629764
Code: github.com/saezlab/greta
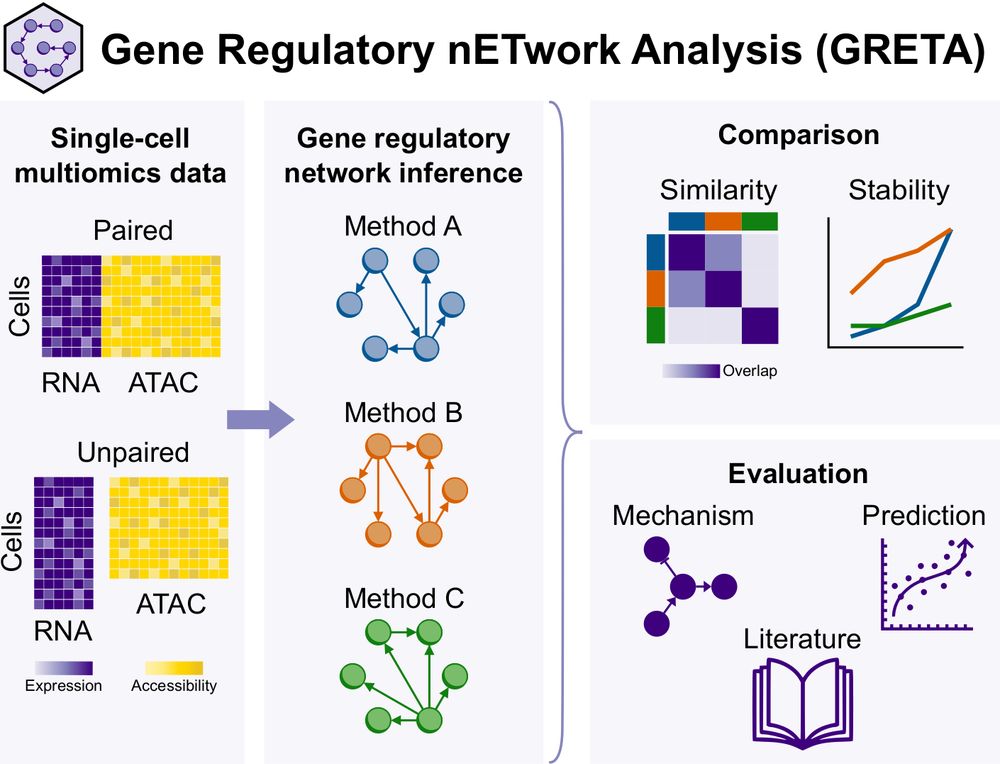
Paper: doi.org/10.1101/2024.12.20.629764
Code: github.com/saezlab/greta
Congratulations to @mariabrbic.bsky.social, Josie Hughues
@trackingactions.bsky.social and Philippe Schwaller! 🥳

Congratulations to @mariabrbic.bsky.social, Josie Hughues
@trackingactions.bsky.social and Philippe Schwaller! 🥳
@EPFL through the #EDIC Program!
📅 Deadline: Dec 15
💥 We're part of School of Computer Science and School of Life Sciences at EPFL which gives access to excellent collaborators, network, and resources!
👉 Learn more at brbiclab.epfl.ch!
@EPFL through the #EDIC Program!
📅 Deadline: Dec 15
💥 We're part of School of Computer Science and School of Life Sciences at EPFL which gives access to excellent collaborators, network, and resources!
👉 Learn more at brbiclab.epfl.ch!

