doi.org/10.1073/pnas...
doi.org/10.1073/pnas...
www.medrxiv.org/content/10.1...

www.medrxiv.org/content/10.1...
Details and instructions for installing on the BEAST website: beast.community

Details and instructions for installing on the BEAST website: beast.community

🔗 doi.org/10.1093/molbev/msaf130
#evobio #molbio #virus
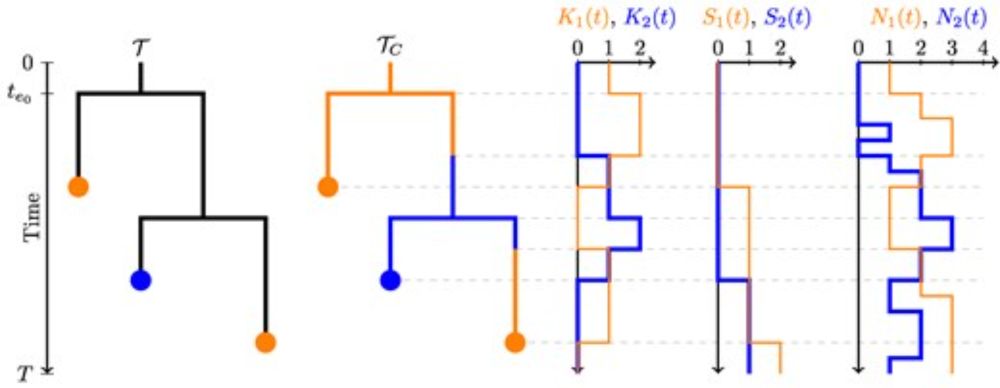
🔗 doi.org/10.1093/molbev/msaf130
#evobio #molbio #virus
📄 tinyurl.com/mt3sffb
🧵 1/6
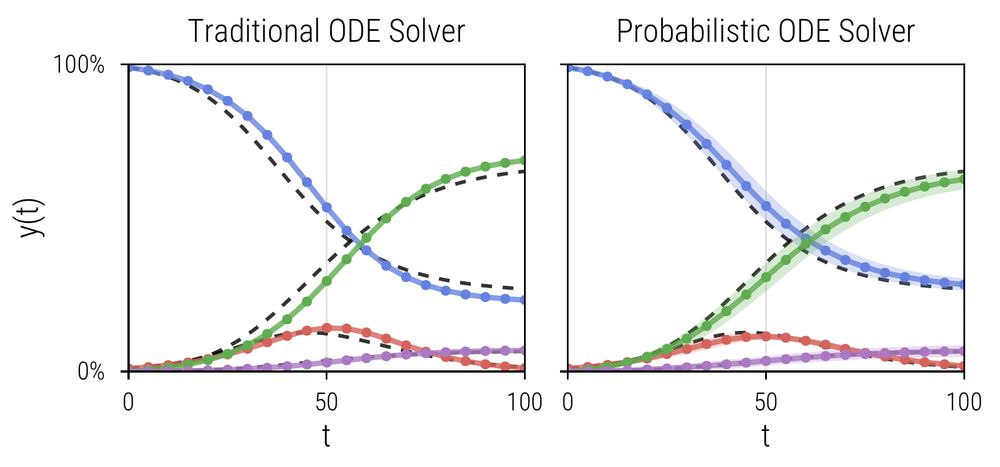
📄 tinyurl.com/mt3sffb
🧵 1/6
1/n
1/n

1/n
🔗 doi.org/10.1093/molb...
#evobio #molbio

🔗 doi.org/10.1093/molb...
#evobio #molbio

🔹 Bern – 31/3 15:15
🔹 Basel – 16/4, 11:15
💡 Talk: Narrow AI for Phylodynamics
This visit is supported by SIB’s Speaker Roadshow Initiative, which coordinates multiple talks at partner institutions and promotes sustainable travel.

🔹 Bern – 31/3 15:15
🔹 Basel – 16/4, 11:15
💡 Talk: Narrow AI for Phylodynamics
This visit is supported by SIB’s Speaker Roadshow Initiative, which coordinates multiple talks at partner institutions and promotes sustainable travel.

