https://gonzalobenegas.github.io/
www.biorxiv.org/content/10.1...
(1/n)

www.biorxiv.org/content/10.1...
(1/n)
1/n

1/n

doi.org/10.1101/2025...
1/n
doi.org/10.1101/2025...
1/n
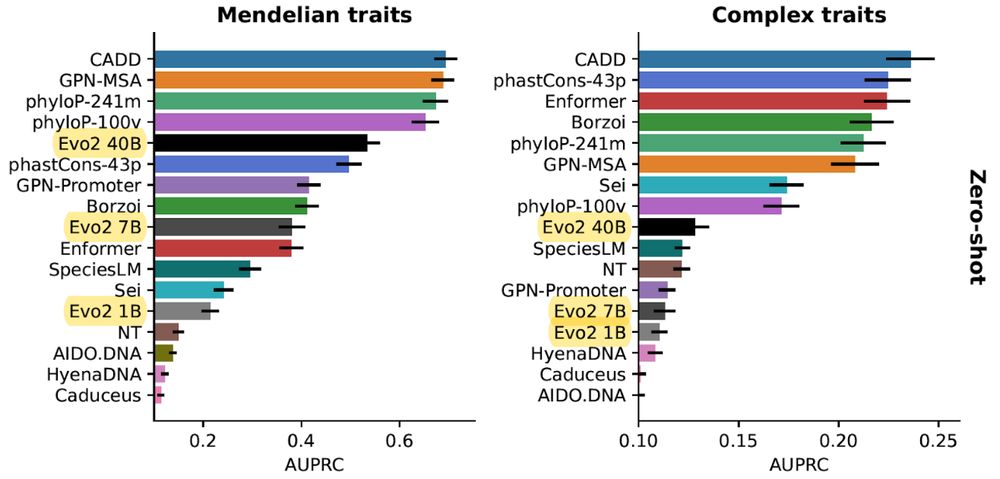
We introduce TraitGym, a curated benchmark of causal regulatory variants for 113 Mendelian & 83 complex traits, and evaluate functional genomics and DNA language models. Joint work w/ Gökcen Eraslan and @yun-s-song.bsky.social 🧵👇

We introduce TraitGym, a curated benchmark of causal regulatory variants for 113 Mendelian & 83 complex traits, and evaluate functional genomics and DNA language models. Joint work w/ Gökcen Eraslan and @yun-s-song.bsky.social 🧵👇
doi.org/10.1093/gene...

doi.org/10.1093/gene...
authors.elsevier.com/a/1kNCscQbJB...
Genomic language models: opportunities and challenges
Please share with your colleagues.
authors.elsevier.com/a/1kNCscQbJB...
Genomic language models: opportunities and challenges
Please share with your colleagues.
doi.org/10.1038/s415...
1/4

doi.org/10.1038/s415...
1/4

1/7
openreview.net/forum?id=H7m...

1/7
openreview.net/forum?id=H7m...

