Thanks to all developers and community members for contributing to the project!
🔗 Please see the full release announcement here: bioconductor.org/news/bioc_3_...

Thanks to all developers and community members for contributing to the project!
🔗 Please see the full release announcement here: bioconductor.org/news/bioc_3_...

We used ddPCA to map the genetic architecture of the entire human bZIP interaction network.
www.biorxiv.org/content/10.1...
Thanks to all our co-authors for the great collaboration!

We used ddPCA to map the genetic architecture of the entire human bZIP interaction network.
www.biorxiv.org/content/10.1...
Thanks to all our co-authors for the great collaboration!
📄 joss.theoj.org/papers/10.2110… (1/3)

📄 joss.theoj.org/papers/10.2110… (1/3)
Submit by July 11 to present your work in Barcelona this September.
🔗 forms.gle/ewTGdKzB2xyE...
📅 Notifications by July 15
#Bioconductor #Bioinformatics
Submit by July 11 to present your work in Barcelona this September.
🔗 forms.gle/ewTGdKzB2xyE...
📅 Notifications by July 15
#Bioconductor #Bioinformatics
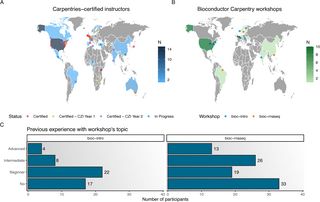
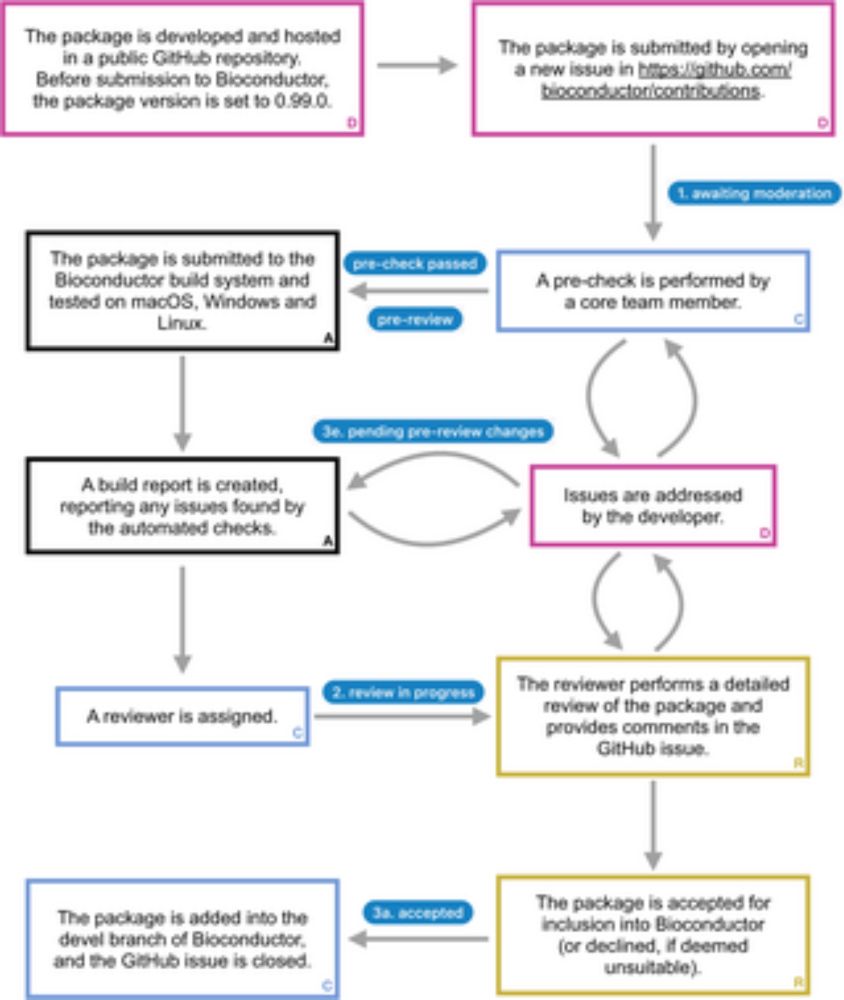
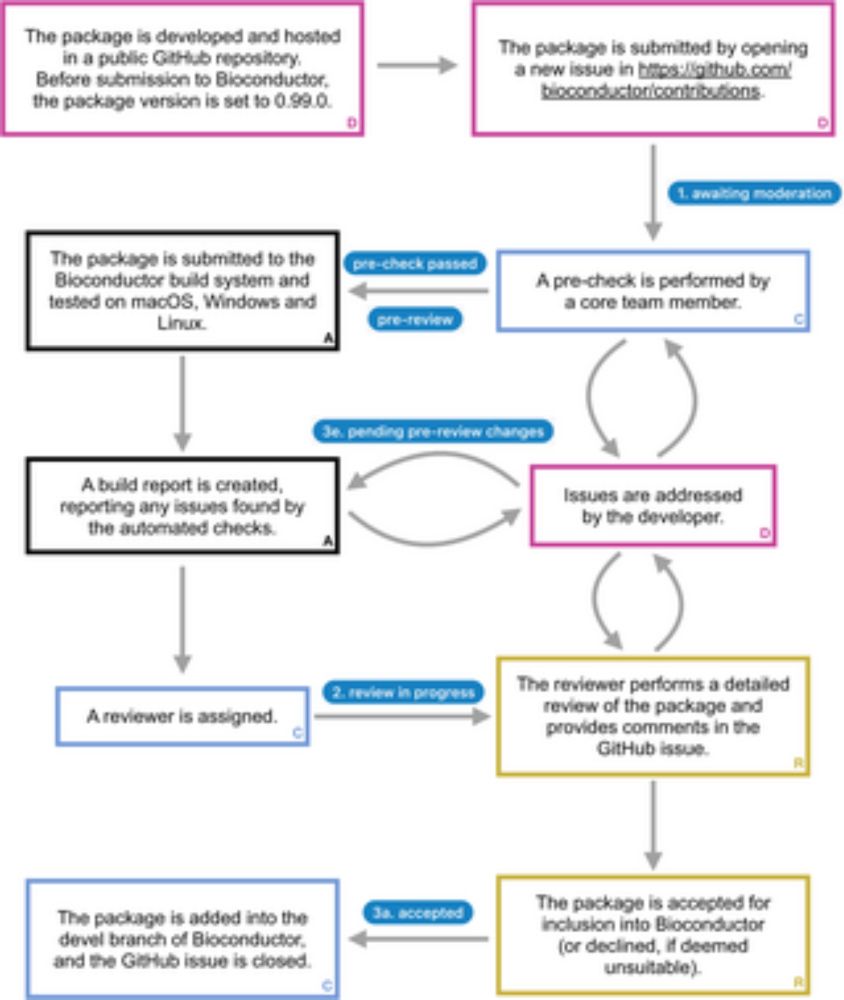
Our 1st paper on Bioconductor training from the Bioconductor Training Committee is now published in PLOS Comp Biol. We're pleased to share our work with the community.
Read here: journals.plos.org/ploscompbiol...
Thank you to everyone involved for your contributions!
Our 1st paper on Bioconductor training from the Bioconductor Training Committee is now published in PLOS Comp Biol. We're pleased to share our work with the community.
Read here: journals.plos.org/ploscompbiol...
Thank you to everyone involved for your contributions!
Join us this Sept in Barcelona to share your research, meet the #Bioconductor community & explore all things #Bioinformatics + #RStats.
👉 eurobioc2025.bioconductor.org/pages/submis...

A huge thank you to all developers and community members for your contributions.
Explore what’s new in the full release announcement:
bioconductor.org/news/bioc_3_...
#Bioconductor #RStats

A huge thank you to all developers and community members for your contributions.
Explore what’s new in the full release announcement:
bioconductor.org/news/bioc_3_...
#Bioconductor #RStats
⬇️
journals.plos.org/ploscompbiol...
⬇️
journals.plos.org/ploscompbiol...
journals.plos.org/ploscompbiol...
journals.plos.org/ploscompbiol...
I am very excited to share our latest work on the protein and chromatin interactomes of transcription factors! 🧬
Huge thanks to everyone who contributed, especially @csoneson.bsky.social and @mbstadler.bsky.social for driving this story with me! 🙌
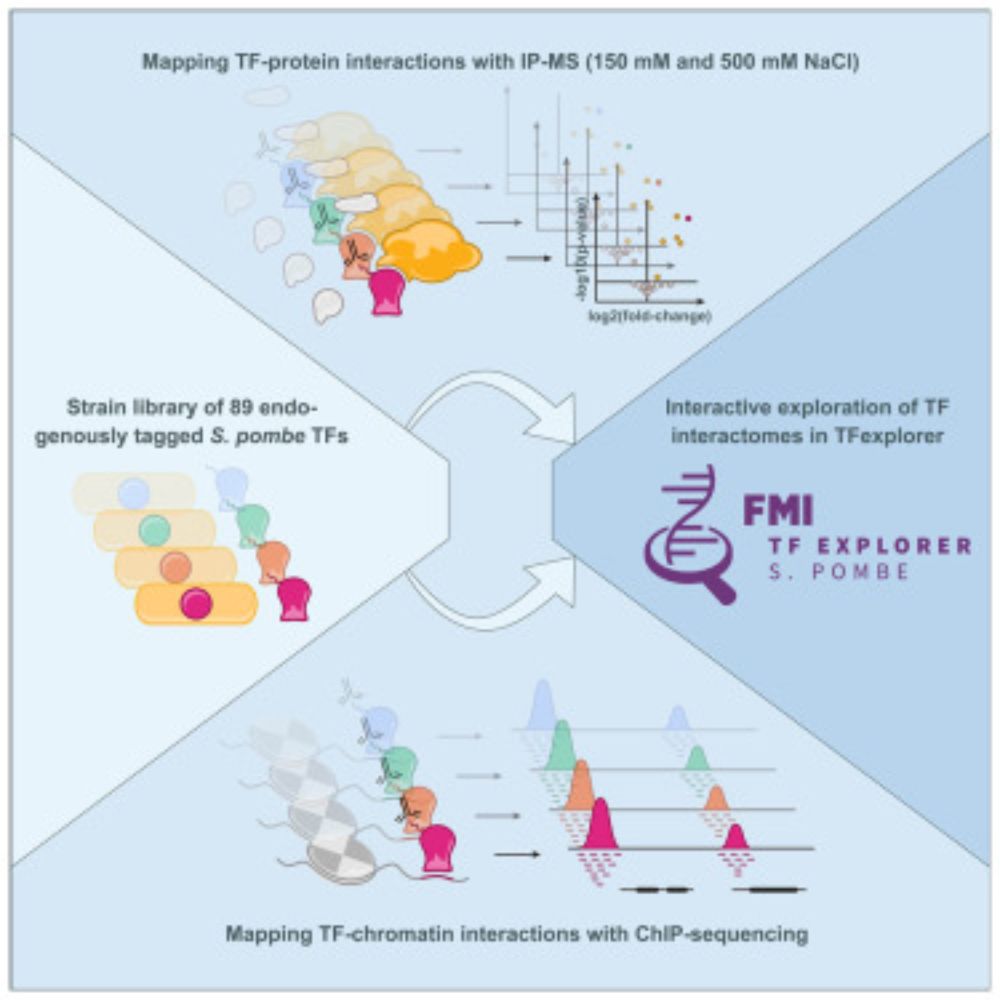
I am very excited to share our latest work on the protein and chromatin interactomes of transcription factors! 🧬
Huge thanks to everyone who contributed, especially @csoneson.bsky.social and @mbstadler.bsky.social for driving this story with me! 🙌

I am very excited to share our latest work on the protein and chromatin interactomes of transcription factors! 🧬
Huge thanks to everyone who contributed, especially @csoneson.bsky.social and @mbstadler.bsky.social for driving this story with me! 🙌


32 researchers | 26 institutes | 10 countries | 4 continents
A #crowd/#community-research success
nature.com/articles/s41...
#CZI, Thanks @mikelove.bsky.social !
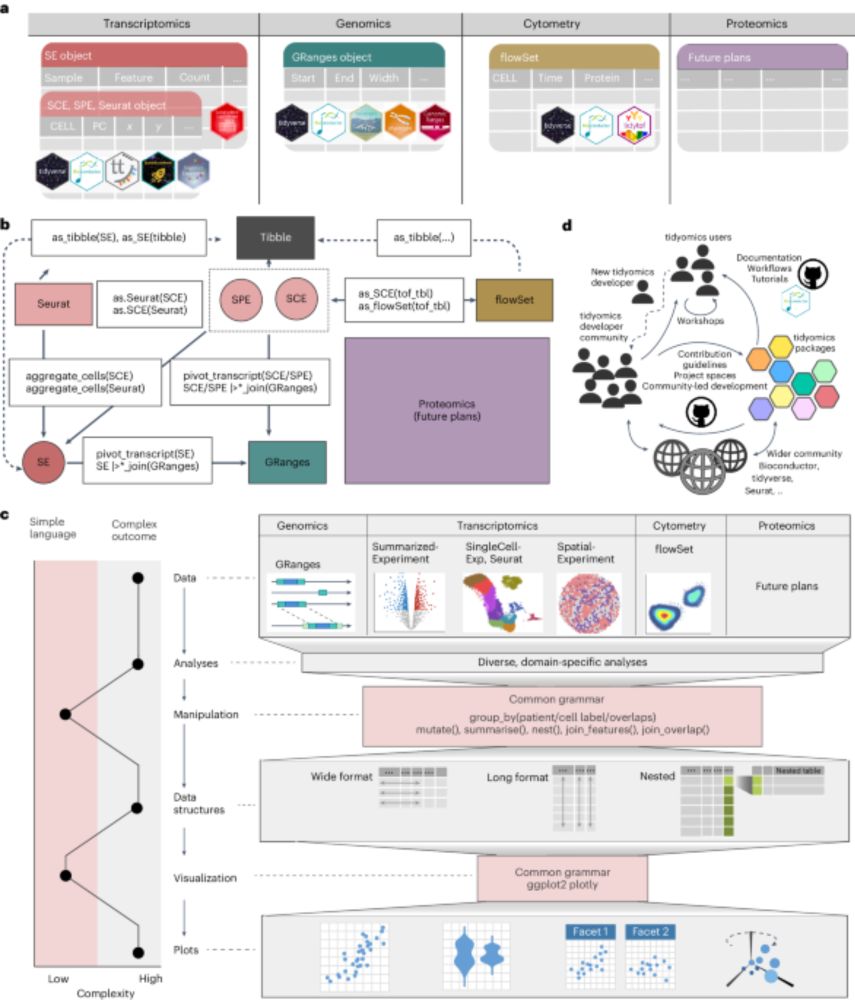
32 researchers | 26 institutes | 10 countries | 4 continents
A #crowd/#community-research success
nature.com/articles/s41...
#CZI, Thanks @mikelove.bsky.social !
📽️ youtu.be/i5I_mFOsGks
📓 docs.google.com/document/d/1...
#rstats #Bioconductor #RNAseq #datavis @federicomarini.bsky.social @csoneson.bsky.social

📽️ youtu.be/i5I_mFOsGks
📓 docs.google.com/document/d/1...
#rstats #Bioconductor #RNAseq #datavis @federicomarini.bsky.social @csoneson.bsky.social

