
Working on proteomics, phosphoproteomics, palaeoproteomics 🧫🔬🦷
We present our collaborative project on “personalising clinical decisions in ovarian cancer through patient-derived in vitro models”.
Have a look at it 👇🏼 and don’t forget to suport us by putting like on Youtube 👍🏼
youtu.be/J4bc1MPqIqE?...

We present our collaborative project on “personalising clinical decisions in ovarian cancer through patient-derived in vitro models”.
Have a look at it 👇🏼 and don’t forget to suport us by putting like on Youtube 👍🏼
youtu.be/J4bc1MPqIqE?...
Read how we did it in our latest paper 👇
pubs.acs.org/doi/10.1021/...
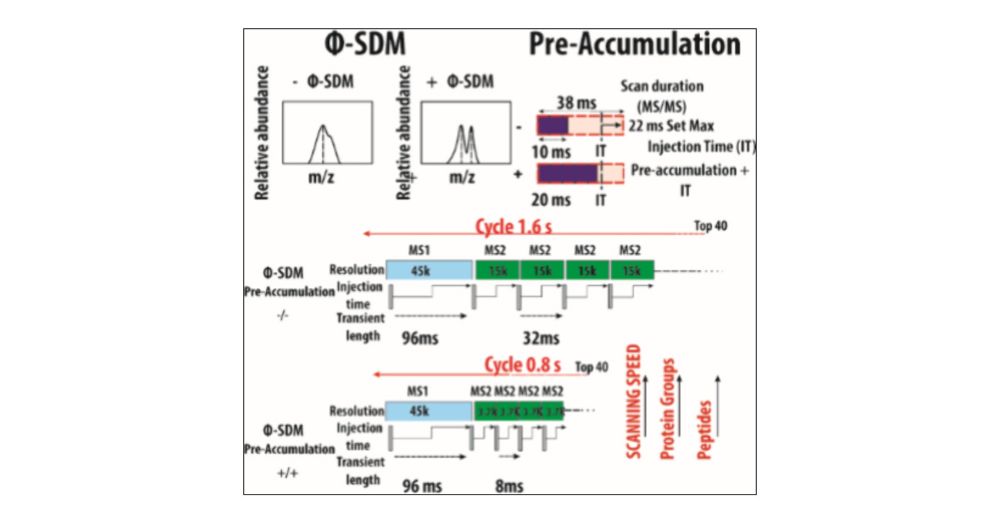
Read how we did it in our latest paper 👇
pubs.acs.org/doi/10.1021/...

We used proteomics on Paranthropus robustus fossils (~2 million years old) to determine the sex of individuals and detect genetic variability, offering new insight into potential subgroups within this ancient hominin 🦷
Our paper on enamel proteins from Paranthropus robustus has finally been peer reviewed, please have a read here: www.science.org/doi/10.1126/...
Paranthropus robustus has been puzzling scientists since its discovery in 1938 in South Africa, where a high number of fossils have been found.
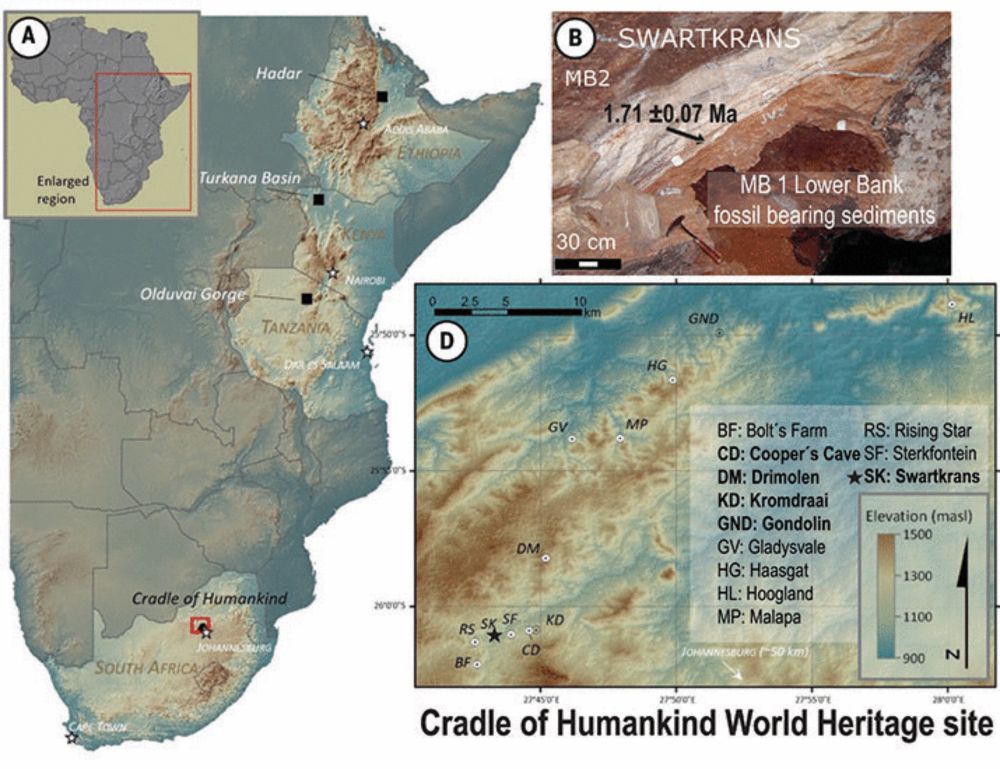
We used proteomics on Paranthropus robustus fossils (~2 million years old) to determine the sex of individuals and detect genetic variability, offering new insight into potential subgroups within this ancient hominin 🦷
Over the years, I’ve gotten multiple questions on the usefulness of ancient proteins in resolving phylogenetic questions. This is our attempt to provide some answers:
www.biorxiv.org/content/10.1...

Over the years, I’ve gotten multiple questions on the usefulness of ancient proteins in resolving phylogenetic questions. This is our attempt to provide some answers:
www.biorxiv.org/content/10.1...
We've developed SC-pSILAC to simultaneously measure protein turnover and abundance in single cells, unlocking the first large-scale, 2D proteomic insights at single-cell resolution!
www.cell.com/cell/fulltex...
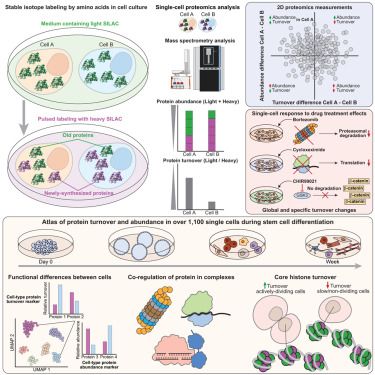
We've developed SC-pSILAC to simultaneously measure protein turnover and abundance in single cells, unlocking the first large-scale, 2D proteomic insights at single-cell resolution!
www.cell.com/cell/fulltex...
We contributed towards the South African Journal of Science Taung Centennial!
some links;
the special issue: issuu.com/sajs/docs/so...
our paper: sajs.co.za/article/view...
and I got featured on where I work from Nature Africa if you are interested in that: www.nature.com/articles/d44...


We contributed towards the South African Journal of Science Taung Centennial!
some links;
the special issue: issuu.com/sajs/docs/so...
our paper: sajs.co.za/article/view...
and I got featured on where I work from Nature Africa if you are interested in that: www.nature.com/articles/d44...
We evaluated how formaldehyde-based fixation preserve proteome state, drug response and cell integrity. Ultimately, cell fixation can facilitate access to #singlecell by enabeling sample shiping and prolonged sorting 📦
pubs.acs.org/doi/10.1021/...
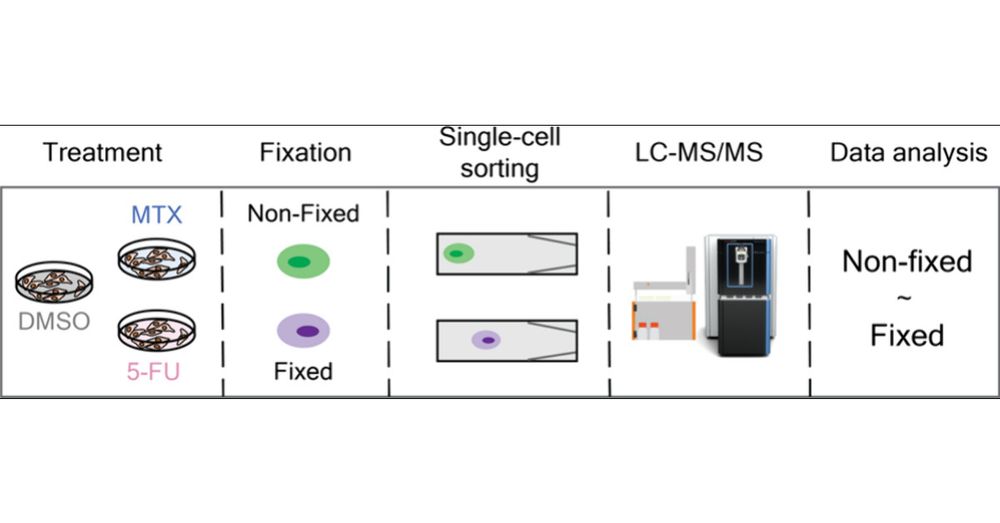
We evaluated how formaldehyde-based fixation preserve proteome state, drug response and cell integrity. Ultimately, cell fixation can facilitate access to #singlecell by enabeling sample shiping and prolonged sorting 📦
pubs.acs.org/doi/10.1021/...
www.nature.com/articles/s41...

www.nature.com/articles/s41...

