



Learn more: https://iwc.galaxyproject.org
Watch: https://youtu.be/cBHSYrpMV8s
##Galaxy (1/2)

Learn more: https://iwc.galaxyproject.org
Watch: https://youtu.be/cBHSYrpMV8s
##Galaxy (1/2)


Name tags make it easy to track dataset origins and relationships across analyses—no endless renaming needed https://galaxyproject.org/news/2025-11-4-tags/
##GalaxyProject ##Bioinformatics ##Reproducibility

Name tags make it easy to track dataset origins and relationships across analyses—no endless renaming needed https://galaxyproject.org/news/2025-11-4-tags/
##GalaxyProject ##Bioinformatics ##Reproducibility
Get ready to join us in Clermont-Ferrand, France for #GCC2026, June 22–26 2026! Learn, collaborate, and celebrate open science in the heart of Auvergne.
galaxyproject.org/news/2025-10...

Get ready to join us in Clermont-Ferrand, France for #GCC2026, June 22–26 2026! Learn, collaborate, and celebrate open science in the heart of Auvergne.
galaxyproject.org/news/2025-10...

Galaxy will be in San Diego for the International Plant & Animal Genome Conference (Jan 9–14, 2026), and we’d love to know who from the Galaxy Community will be there!
Let us know 👉 forms.gle/26PRwMH46aho...

Galaxy will be in San Diego for the International Plant & Animal Genome Conference (Jan 9–14, 2026), and we’d love to know who from the Galaxy Community will be there!
Let us know 👉 forms.gle/26PRwMH46aho...

Check out the blog (https://galaxyproject.org/news/2025-10-21-whycollections/) and video (https://youtu.be/yehY9tR_NX8?si=YH9CZ5KGWm2yVyPA) to see how they can simplify your workflows.
You might just find yourself wondering, (1/2)
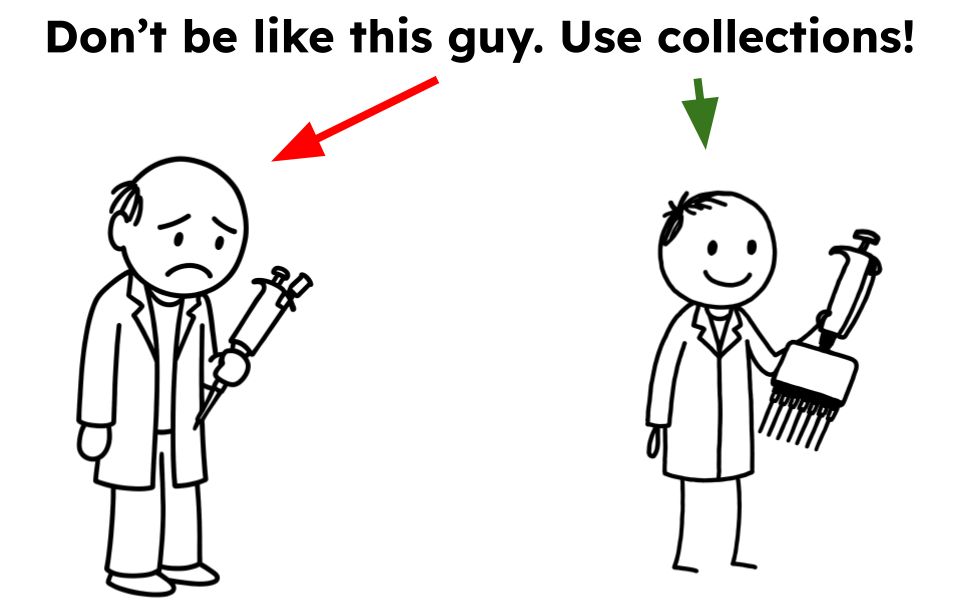
Check out the blog (https://galaxyproject.org/news/2025-10-21-whycollections/) and video (https://youtu.be/yehY9tR_NX8?si=YH9CZ5KGWm2yVyPA) to see how they can simplify your workflows.
You might just find yourself wondering, (1/2)
Presenters introduced the three BRCs (BV-BRC, PDN and BRC-Analytics) in a tight 30 minutes.
@bvbrc.bsky.social

Presenters introduced the three BRCs (BV-BRC, PDN and BRC-Analytics) in a tight 30 minutes.
@bvbrc.bsky.social

https://gxy.io/brc-webinar-reg
@bvbrc.bsky.social

https://gxy.io/brc-webinar-reg
@bvbrc.bsky.social
https://gxy.io/brc-webinar-reg
@bvbrc.bsky.social

https://gxy.io/brc-webinar-reg
@bvbrc.bsky.social
Register: https://www.bv-brc.org/brc-niaid-ai-codeathon-2025
##UseGalaxy

Register: https://www.bv-brc.org/brc-niaid-ai-codeathon-2025
##UseGalaxy
Register here: https://events.teams.microsoft.com/event/26e362e7-0d72-4c6a-9e86-0fd306368dfd@24d967f1-3ed8-4448-baa6-560ec572acb3
##UseGalaxy ##BVBRC

Register here: https://events.teams.microsoft.com/event/26e362e7-0d72-4c6a-9e86-0fd306368dfd@24d967f1-3ed8-4448-baa6-560ec572acb3
##UseGalaxy ##BVBRC

https://youtu.be/lYzb0j_UtWk
Please follow for more updates and to give feedback on https://BRC-Analytics.org
##UseGalaxy

https://youtu.be/lYzb0j_UtWk
Please follow for more updates and to give feedback on https://BRC-Analytics.org
##UseGalaxy
See (1/2)

See (1/2)

https://galaxyproject.org/events/2025-03-26-brc-galaxy-webinar/
for more information.
##UseGalaxy ##NIAID ##BRC

https://galaxyproject.org/events/2025-03-26-brc-galaxy-webinar/
for more information.
##UseGalaxy ##NIAID ##BRC

