http://jozhang97.github.io
🔖 paper www.biorxiv.org/content/10.1...
💻 github: github.com/jozhang97/ISM
🤗 huggingface: huggingface.co/jozhang97/is...
🔖 paper www.biorxiv.org/content/10.1...
💻 github: github.com/jozhang97/ISM
🤗 huggingface: huggingface.co/jozhang97/is...
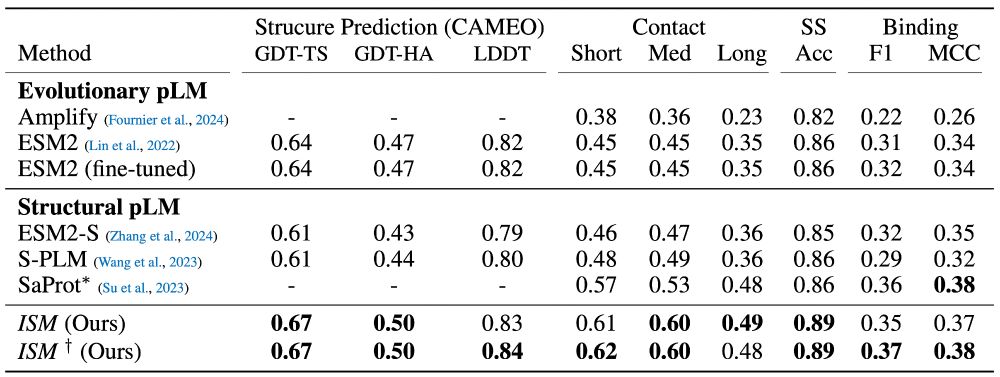
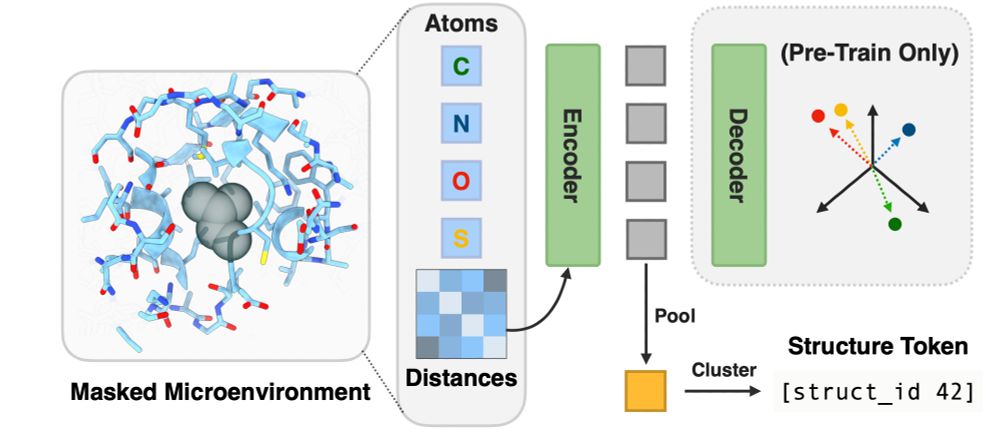
We fine-tune ESM2 to predict representations from structure models. (3/7)
We fine-tune ESM2 to predict representations from structure models. (3/7)

