
Group Leader of @ccg-ucl.bsky.social at the UCL Cancer Institute, @CRUKLungCentre, and Visiting Scientist at the Francis Crick
W: http://www.ucl.ac.uk/cancer/zaccaria-lab
W: https://sites.google.com/view/ccgresearchgroup
@cancerresearchuk.org 's blog post 👇
news.cancerresearchuk.org/2024/02/27/u...

@cancerresearchuk.org 's blog post 👇
news.cancerresearchuk.org/2024/02/27/u...
🖥️SPRINTER is fully available in GitHub 👉 github.com/zaccaria-lab...
🚆Distributed through Bioconda (with also related container) 👉 bioconda.github.io/recipes/spri...
💾also with a reproducible capsule in CodeOcean👉 doi.org/10.24433/CO.... pic.x.com/zpL2rhbRhl
🖥️SPRINTER is fully available in GitHub 👉 github.com/zaccaria-lab...
🚆Distributed through Bioconda (with also related container) 👉 bioconda.github.io/recipes/spri...
💾also with a reproducible capsule in CodeOcean👉 doi.org/10.24433/CO.... pic.x.com/zpL2rhbRhl
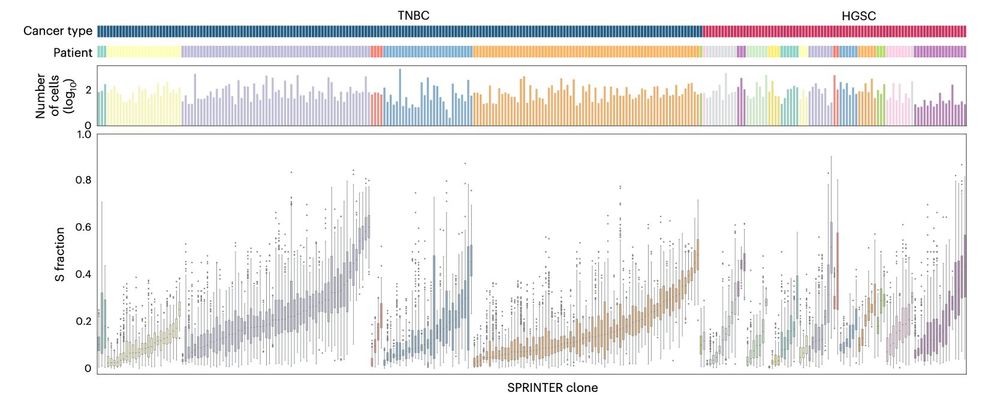
❗️Increased single-cell rates of genomic variants
‼️Enrichment of prolif-related gene amplifications
❔-> Evolutionary advantage?
❗️Increased single-cell rates of genomic variants
‼️Enrichment of prolif-related gene amplifications
❔-> Evolutionary advantage?
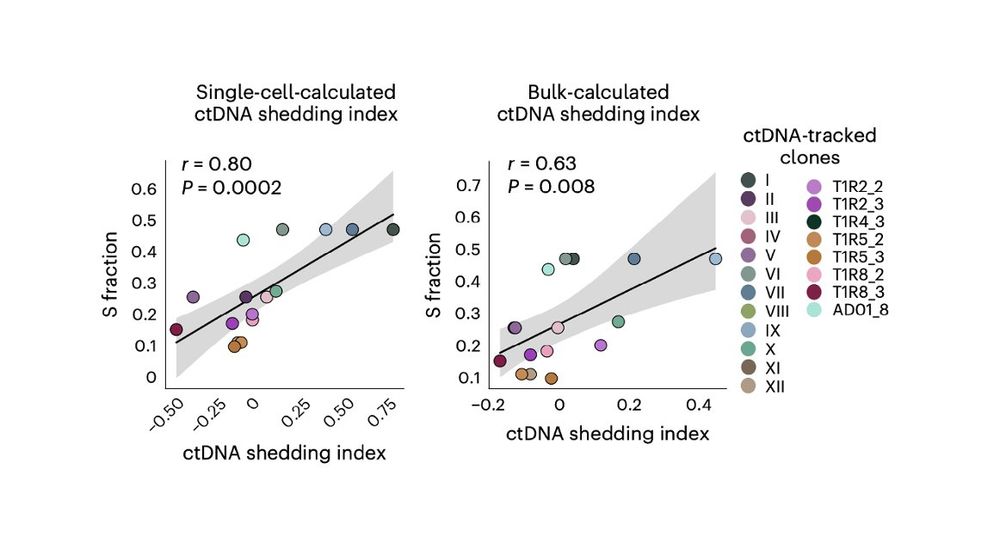
🚨It shows that previous similar observations obtained across different tumors also hold for distinct clones within a tumor
🚨It shows that previous similar observations obtained across different tumors also hold for distinct clones within a tumor
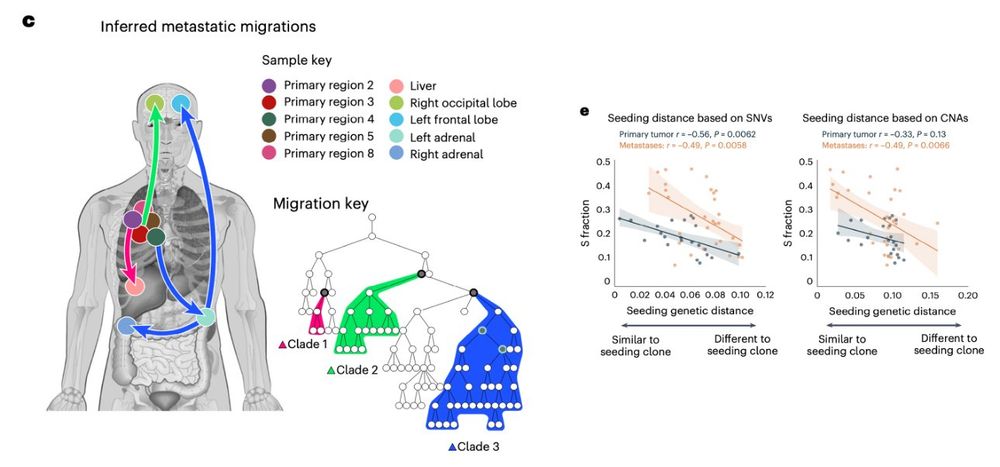
1. high prolif clones seeded most mets disseminating from other mets
2. met-seeding clones had high proliferation
❔-> association between high prolif and met seeding potential of individual clones
1. high prolif clones seeded most mets disseminating from other mets
2. met-seeding clones had high proliferation
❔-> association between high prolif and met seeding potential of individual clones
‼️ We found ART unique to the high prolif and metastatic clones affecting key genes like KRAS, and with associated expression changes from matched RNA-seq
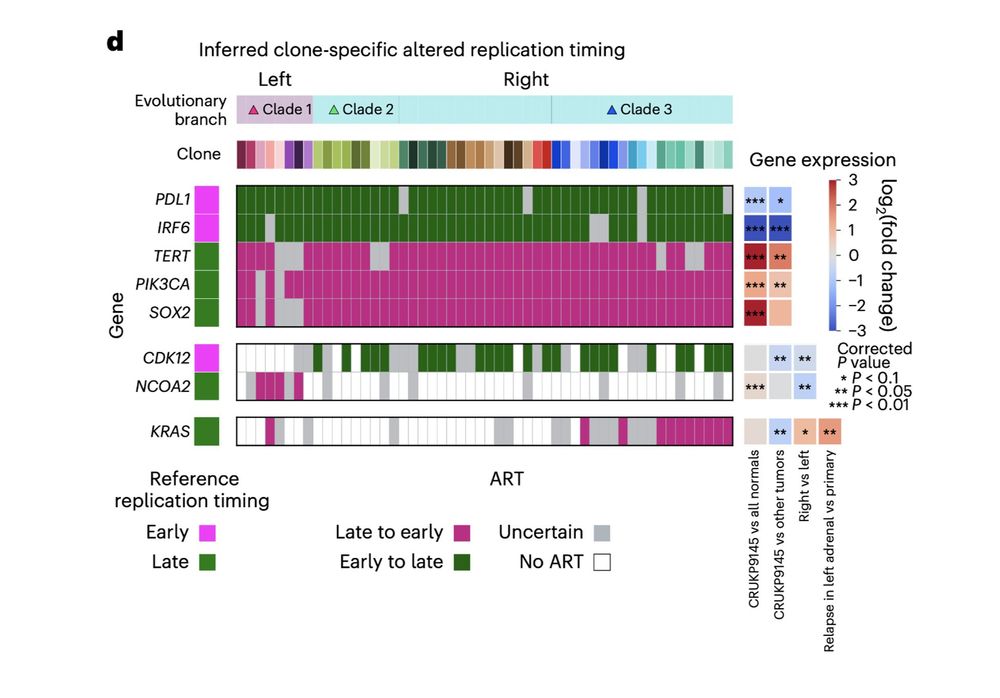
‼️ We found ART unique to the high prolif and metastatic clones affecting key genes like KRAS, and with associated expression changes from matched RNA-seq
⛔️ No clearly associated genetic driver was found
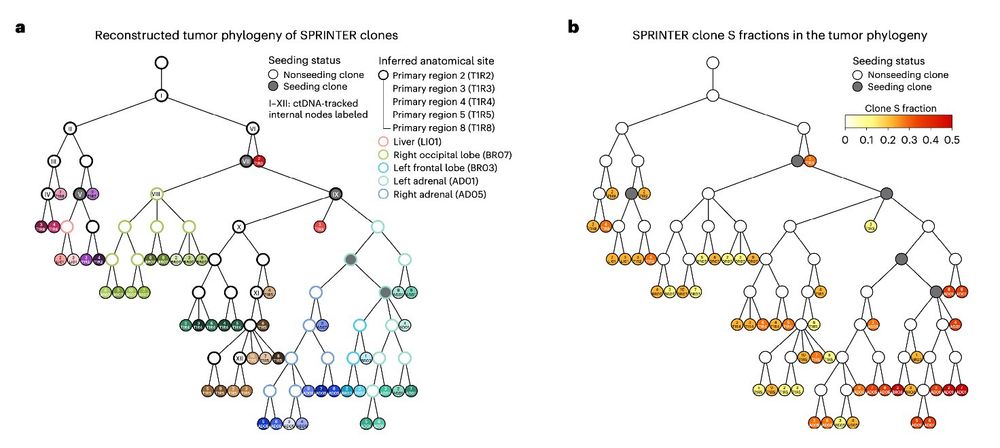
⛔️ No clearly associated genetic driver was found
❓-> Yes, clones can proliferate differently
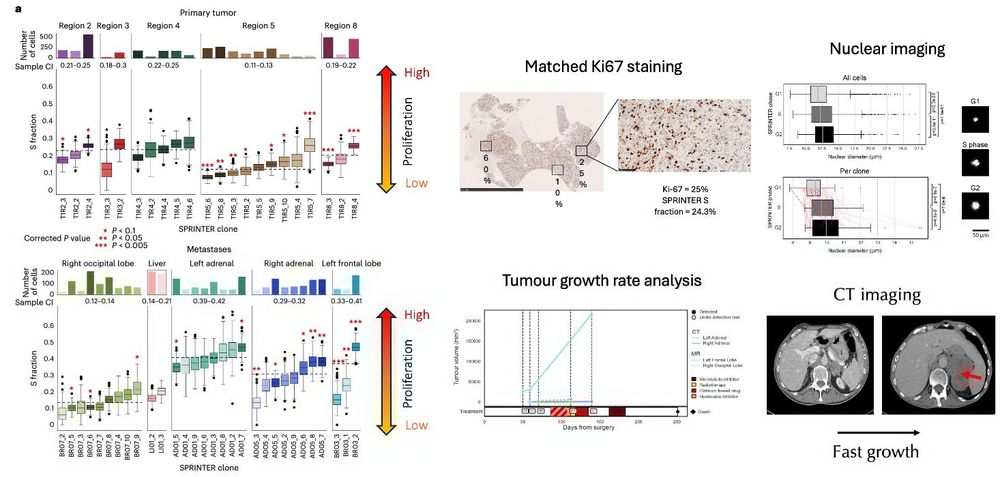
❓-> Yes, clones can proliferate differently
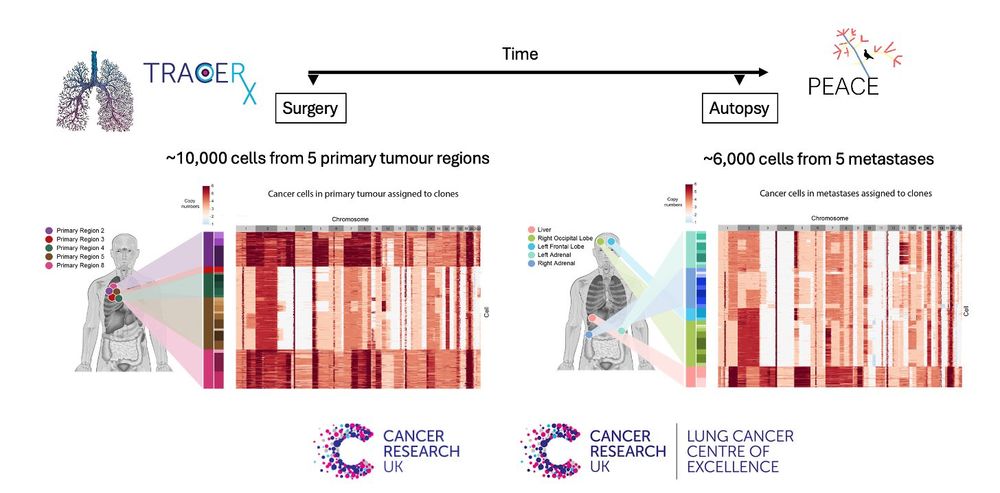




⚠️ But estimating clone proliferation remains challenging due to lack of formal methods for clone assignment of replicating cells and low identification power

⚠️ But estimating clone proliferation remains challenging due to lack of formal methods for clone assignment of replicating cells and low identification power
❓Can it also differ between distinct clones co-existing within a tumor?
❔If so, do proliferation differences associate with more aggressive phenotypes or other properties of these clones?

❓Can it also differ between distinct clones co-existing within a tumor?
❔If so, do proliferation differences associate with more aggressive phenotypes or other properties of these clones?

