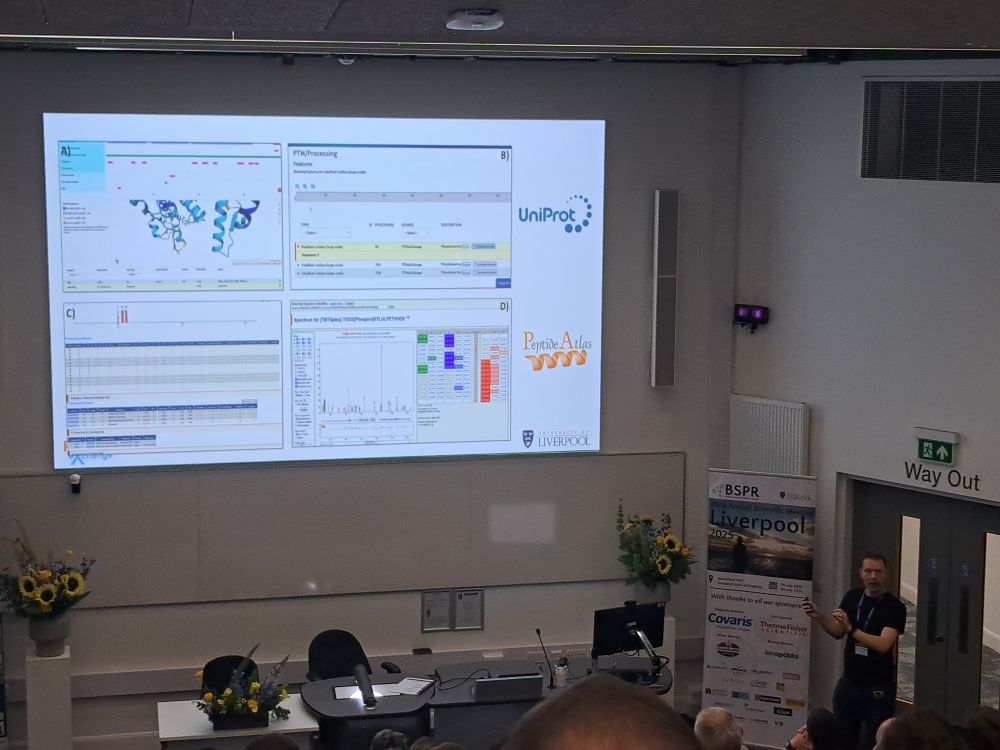
omics, data science, and coding
Lead of bigbio projects



💡 Create stunning, shareable HTML reports for your collaborators in seconds.
✨ Try pmultiqc.quantms.org Examples👇
#Proteomics #QC
💡 Create stunning, shareable HTML reports for your collaborators in seconds.
✨ Try pmultiqc.quantms.org Examples👇
#Proteomics #QC

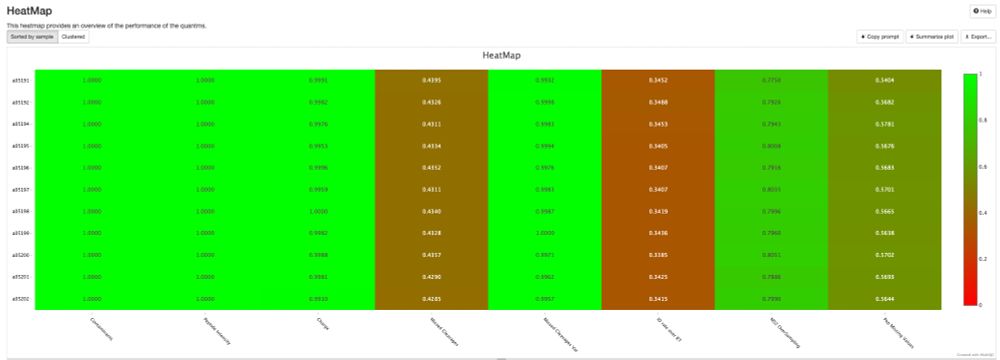
Here are the statistics of his #HipSci dataset, which has remained at 2% after 6 years. 📈
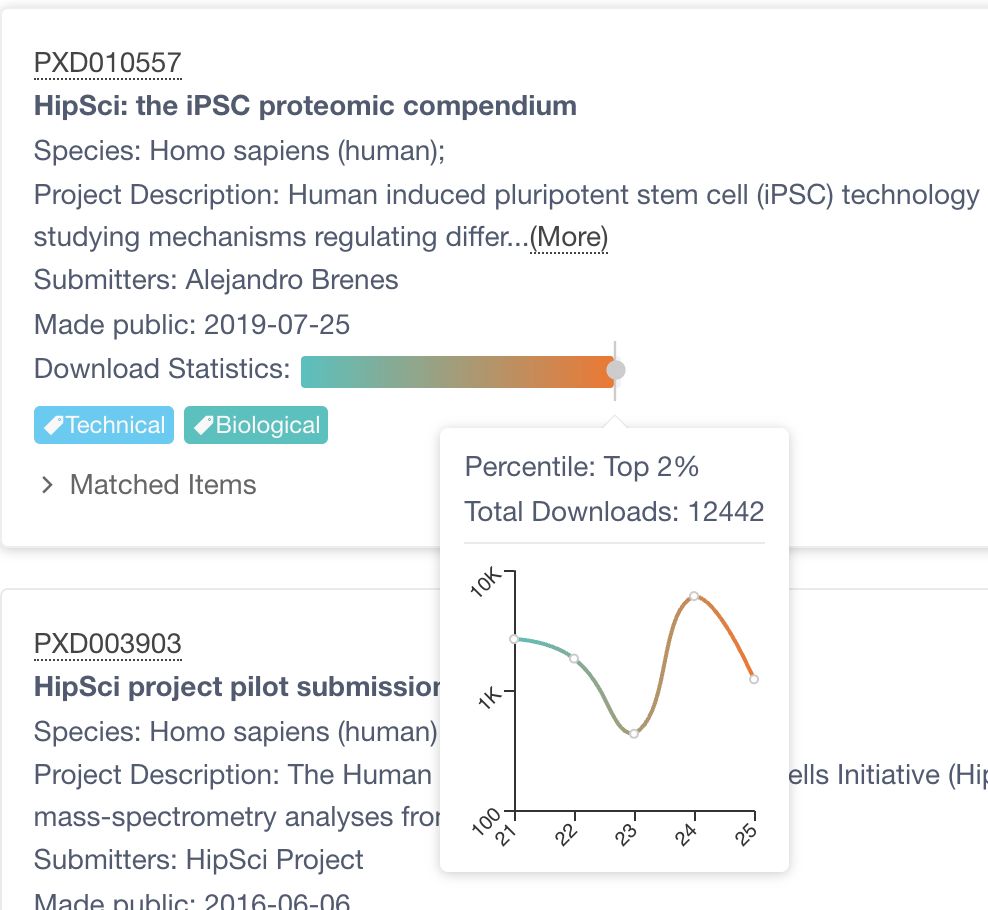
Here are the statistics of his #HipSci dataset, which has remained at 2% after 6 years. 📈


Why else would download rates spike like it’s a secret mass spectrometry ritual? 🧙
Im preparing the team for next August. 🧑💻🏝️

Why else would download rates spike like it’s a secret mass spectrometry ritual? 🧙
Im preparing the team for next August. 🧑💻🏝️


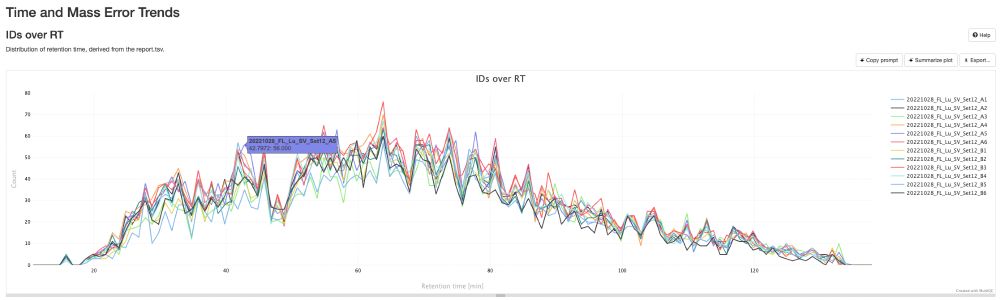
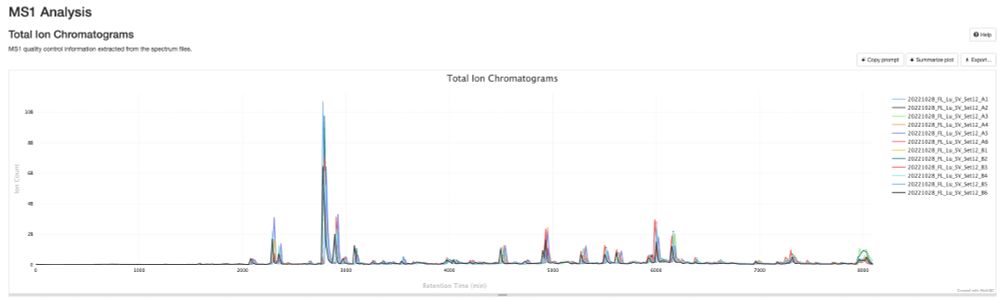
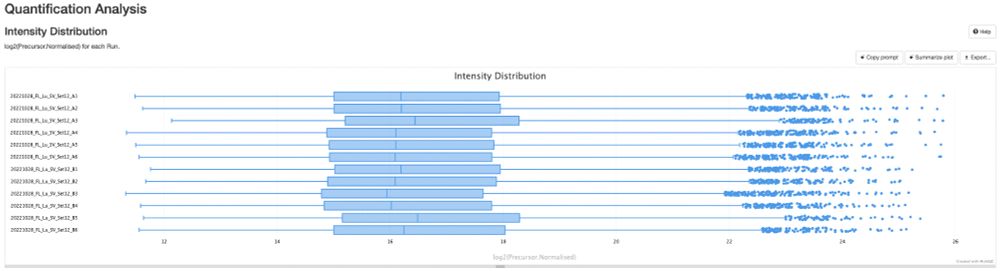
pmultiqc.quantms.org now supports #DIA-NN! 🎉
Just drop in your diann_report.tsv, run quantms-utils, and boom 💥—beautiful summary reports for your spectra! 📊✨
Here an example: pmultiqc.quantms.org/DIANN/multiq...
Check out some sweet plots 👇 #proteomics #massspec


pmultiqc.quantms.org now supports #DIA-NN! 🎉
Just drop in your diann_report.tsv, run quantms-utils, and boom 💥—beautiful summary reports for your spectra! 📊✨
Here an example: pmultiqc.quantms.org/DIANN/multiq...
Check out some sweet plots 👇 #proteomics #massspec