
Kit Curtius
@yosoykit.bsky.social
Assistant Professor at UCSD Medicine, Division of Biomedical Informatics.
Researching cancer evolution and early detection. Check out our work at www.qcclab.com
Researching cancer evolution and early detection. Check out our work at www.qcclab.com
All our methods using open weight LLMs are publicly available to modify for other problems and explained in detail in our paper and SI: github.com/bdj34/llama....
GitHub - bdj34/llama.cpp_data_extraction: Back up code to structure pathology reports. Repository was originally a clone of llama.cpp, then modified for data structuring
Back up code to structure pathology reports. Repository was originally a clone of llama.cpp, then modified for data structuring - bdj34/llama.cpp_data_extraction
github.com
September 19, 2025 at 3:33 PM
All our methods using open weight LLMs are publicly available to modify for other problems and explained in detail in our paper and SI: github.com/bdj34/llama....
thank you for the support Trevor!
August 28, 2025 at 5:11 PM
thank you for the support Trevor!
No worries Suzanne, difficult last name club! haha thank you for the support <3
July 17, 2025 at 3:02 AM
No worries Suzanne, difficult last name club! haha thank you for the support <3
It's open to the public I just walked right on! 7 mins walk from the convention center :)
July 15, 2025 at 8:43 PM
It's open to the public I just walked right on! 7 mins walk from the convention center :)
Will the recording be up on youtube after streaming? Have a meeting at same time but would like to watch :)
February 25, 2025 at 6:40 PM
Will the recording be up on youtube after streaming? Have a meeting at same time but would like to watch :)
Finally, we are very hopeful that this genetic test could help prevent cancers in very high-risk patients. We are pushing this work forward still, much more to come! Thank you to the fantastic team I had the pleasure working with the past years @trevorgraham.bsky.social et al @cancerresearchuk.org
January 30, 2025 at 4:05 PM
Finally, we are very hopeful that this genetic test could help prevent cancers in very high-risk patients. We are pushing this work forward still, much more to come! Thank you to the fantastic team I had the pleasure working with the past years @trevorgraham.bsky.social et al @cancerresearchuk.org
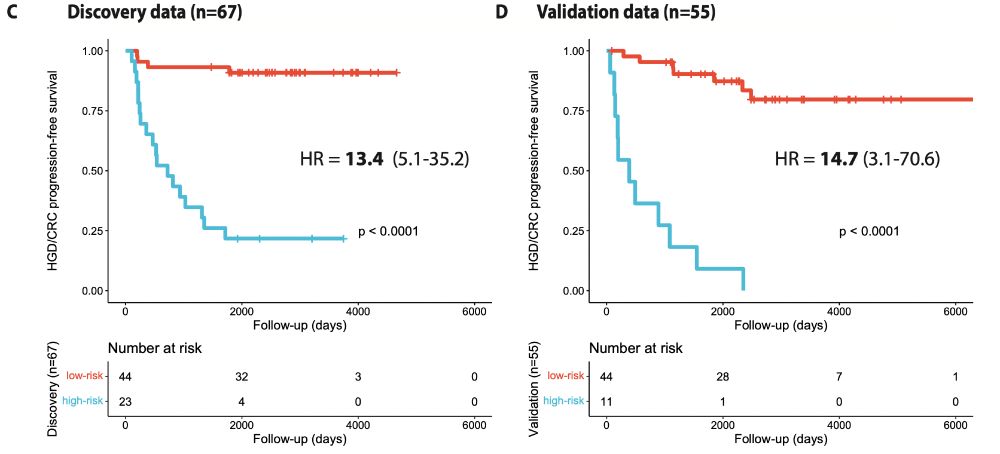
With independent data from other UK centers, we externally validated this biomarker and found similar risk stratification. As a predictive biomarker for HGD/CRC, both PPV & NPV >90% at 5 years post initial LGD resection time point in the validation. Check out the paper for more!
January 30, 2025 at 4:02 PM
With independent data from other UK centers, we externally validated this biomarker and found similar risk stratification. As a predictive biomarker for HGD/CRC, both PPV & NPV >90% at 5 years post initial LGD resection time point in the validation. Check out the paper for more!
We found LGD from progressors already harbored significantly more CNAs than non-progressors at index Dx. As a binary biomarker, the odds ratio of developing HGD/CRC for a high CNA score was 36, and this was 6 times higher than OR for even the strongest clinical predictor (invisible/unresected LGD).

January 30, 2025 at 4:00 PM
We found LGD from progressors already harbored significantly more CNAs than non-progressors at index Dx. As a binary biomarker, the odds ratio of developing HGD/CRC for a high CNA score was 36, and this was 6 times higher than OR for even the strongest clinical predictor (invisible/unresected LGD).
Low-grade dysplasia is notoriously hard to risk-stratify, even with some known risk factors for later cancer (e.g., size, multifocality). We hypothesized that the true culprit was underlying altered genomes of cells and conducted a large case-control study to test this at index LGD timepoint.

January 30, 2025 at 3:58 PM
Low-grade dysplasia is notoriously hard to risk-stratify, even with some known risk factors for later cancer (e.g., size, multifocality). We hypothesized that the true culprit was underlying altered genomes of cells and conducted a large case-control study to test this at index LGD timepoint.
This is just the start of our work to tease apart relationships between host-microbe clonal populations in BE progression from an evolutionary perspective. Thanks for the fun collaboration with labs @ucsdmedschool.bsky.social @ucsandiego.bsky.social @fredhutch.bsky.social !
January 17, 2025 at 4:23 PM
This is just the start of our work to tease apart relationships between host-microbe clonal populations in BE progression from an evolutionary perspective. Thanks for the fun collaboration with labs @ucsdmedschool.bsky.social @ucsandiego.bsky.social @fredhutch.bsky.social !
Finally in model simulations, we found that assuming widely varying growth rates across taxa best reflected EAC data, implying that selection pressures may influence largely niche-based population dynamics in the tumor microenvironment.
January 17, 2025 at 4:18 PM
Finally in model simulations, we found that assuming widely varying growth rates across taxa best reflected EAC data, implying that selection pressures may influence largely niche-based population dynamics in the tumor microenvironment.
We found evidence of mostly neutral processes in normal and precancerous tissue, with the exception of Helicobacter pylori which deviated from a neutral expectation in BE patients who did not progress to EAC. For the EAC microbiome, patterns seem largely non-neutral.
January 17, 2025 at 4:16 PM
We found evidence of mostly neutral processes in normal and precancerous tissue, with the exception of Helicobacter pylori which deviated from a neutral expectation in BE patients who did not progress to EAC. For the EAC microbiome, patterns seem largely non-neutral.

