#MicrobiomeTransmission #OralMicrobiome
https://linktr.ee/vitorheidrich
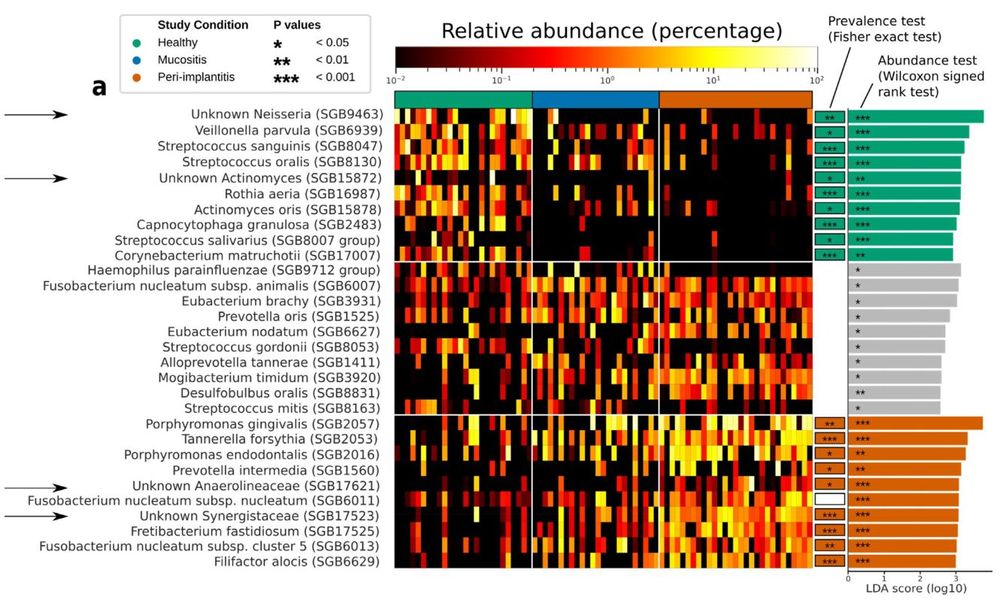
www.nature.com/articles/s41...
Have a nice read!
@cibiocm.bsky.social
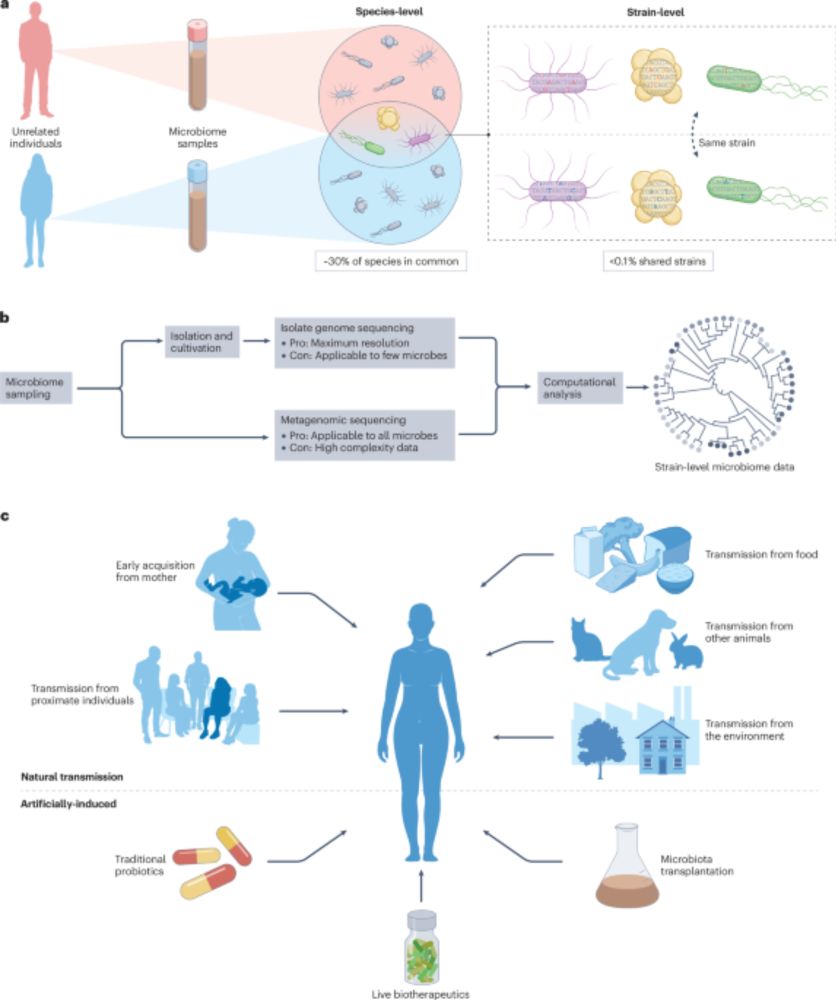
www.nature.com/articles/s41...
Have a nice read!
@cibiocm.bsky.social
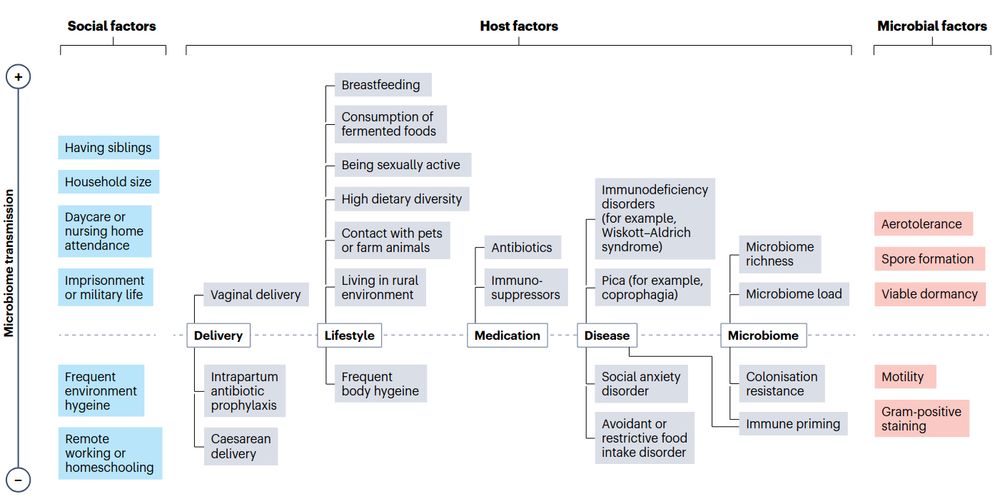
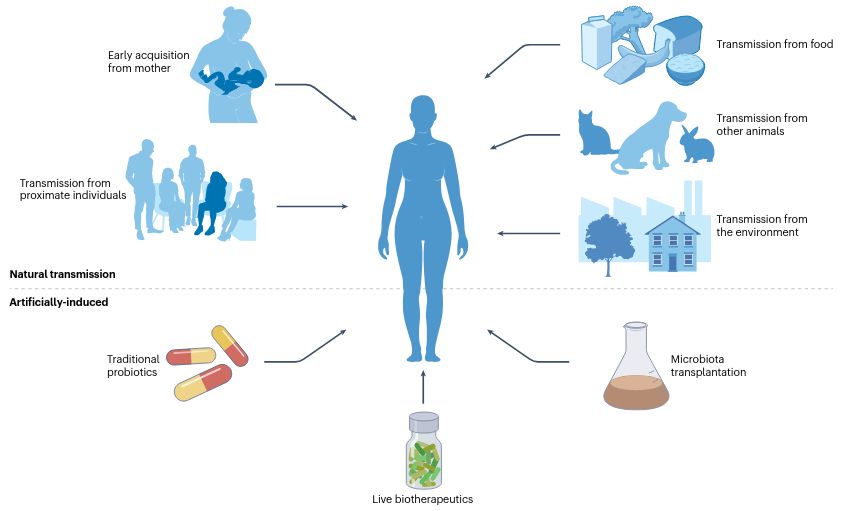
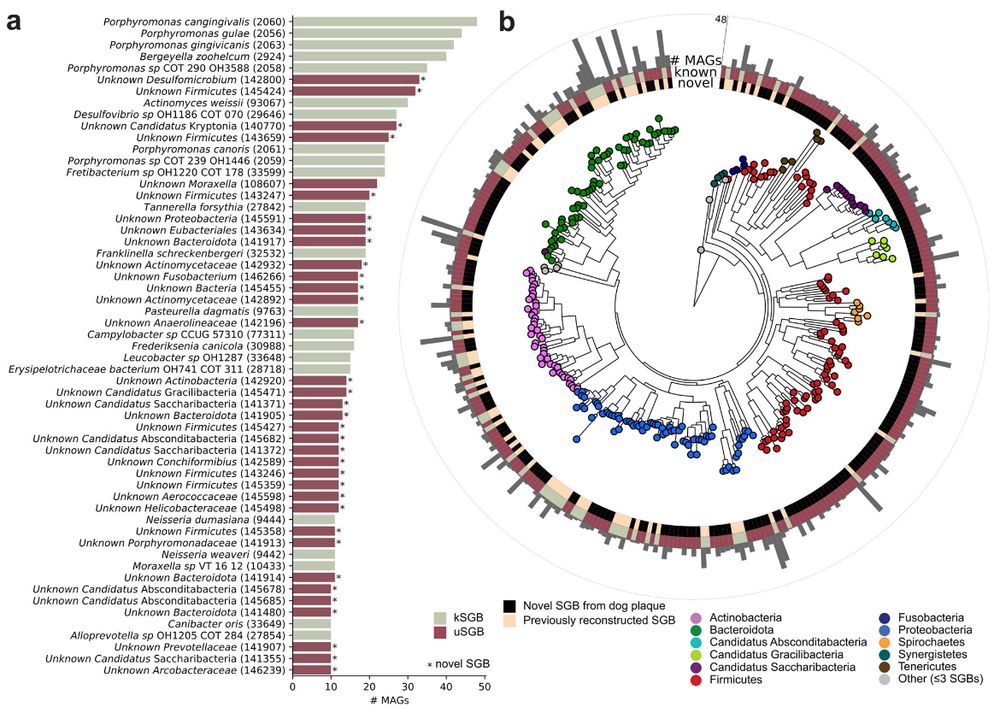
@fackelmeister.bsky.social @raj09.bsky.social @cengoni.bsky.social @cibiocm.bsky.social
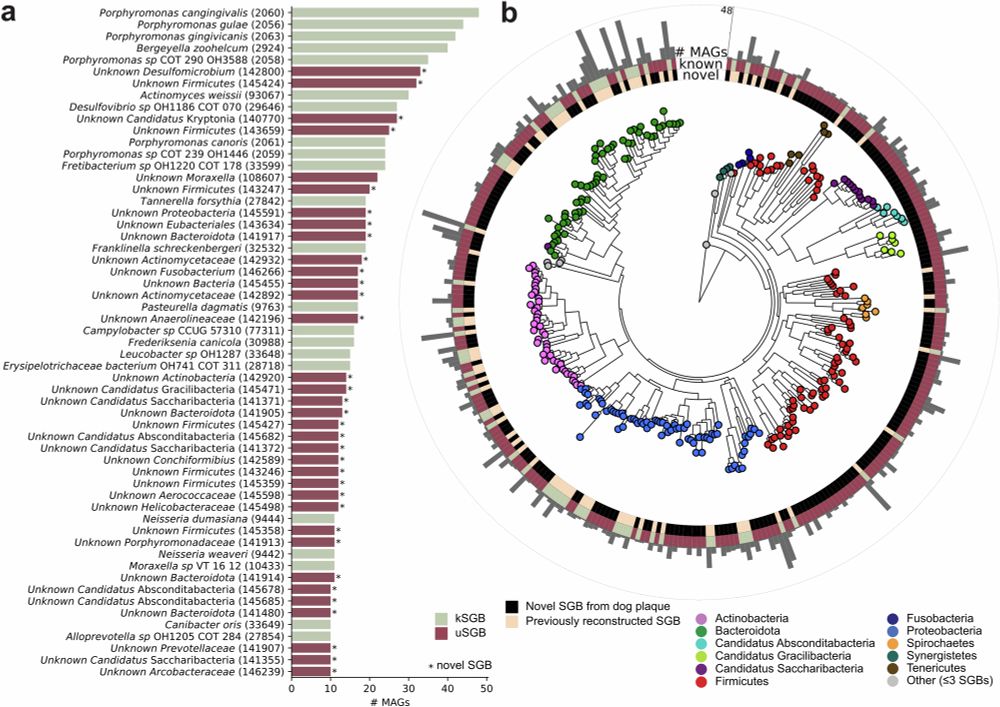
@fackelmeister.bsky.social @raj09.bsky.social @cengoni.bsky.social @cibiocm.bsky.social