Marie-Eve VAL
@val-meve.bsky.social
Microbiologist, plasmid biology, genome maintenance, DNA replication, Vibrio, Institut Pasteur - Paris
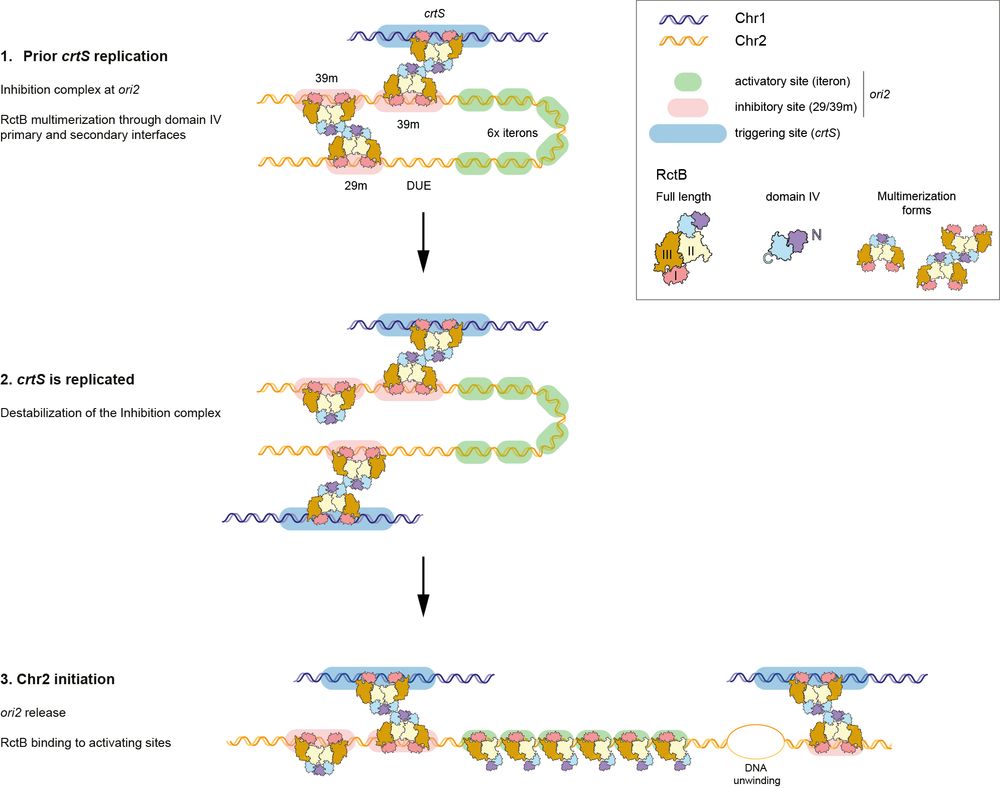
6/ 💡 Proposed model
Before crtS replication: ori2 is OFF, blocked by inhibitory RctB-DNA loops.
After crtS replication: inhibition is destabilized, ori2 becomes accessible, and Chr2 replication begins.
A beautiful molecular ON/OFF switch! 🕹️
Before crtS replication: ori2 is OFF, blocked by inhibitory RctB-DNA loops.
After crtS replication: inhibition is destabilized, ori2 becomes accessible, and Chr2 replication begins.
A beautiful molecular ON/OFF switch! 🕹️
January 9, 2025 at 12:47 PM
6/ 💡 Proposed model
Before crtS replication: ori2 is OFF, blocked by inhibitory RctB-DNA loops.
After crtS replication: inhibition is destabilized, ori2 becomes accessible, and Chr2 replication begins.
A beautiful molecular ON/OFF switch! 🕹️
Before crtS replication: ori2 is OFF, blocked by inhibitory RctB-DNA loops.
After crtS replication: inhibition is destabilized, ori2 becomes accessible, and Chr2 replication begins.
A beautiful molecular ON/OFF switch! 🕹️
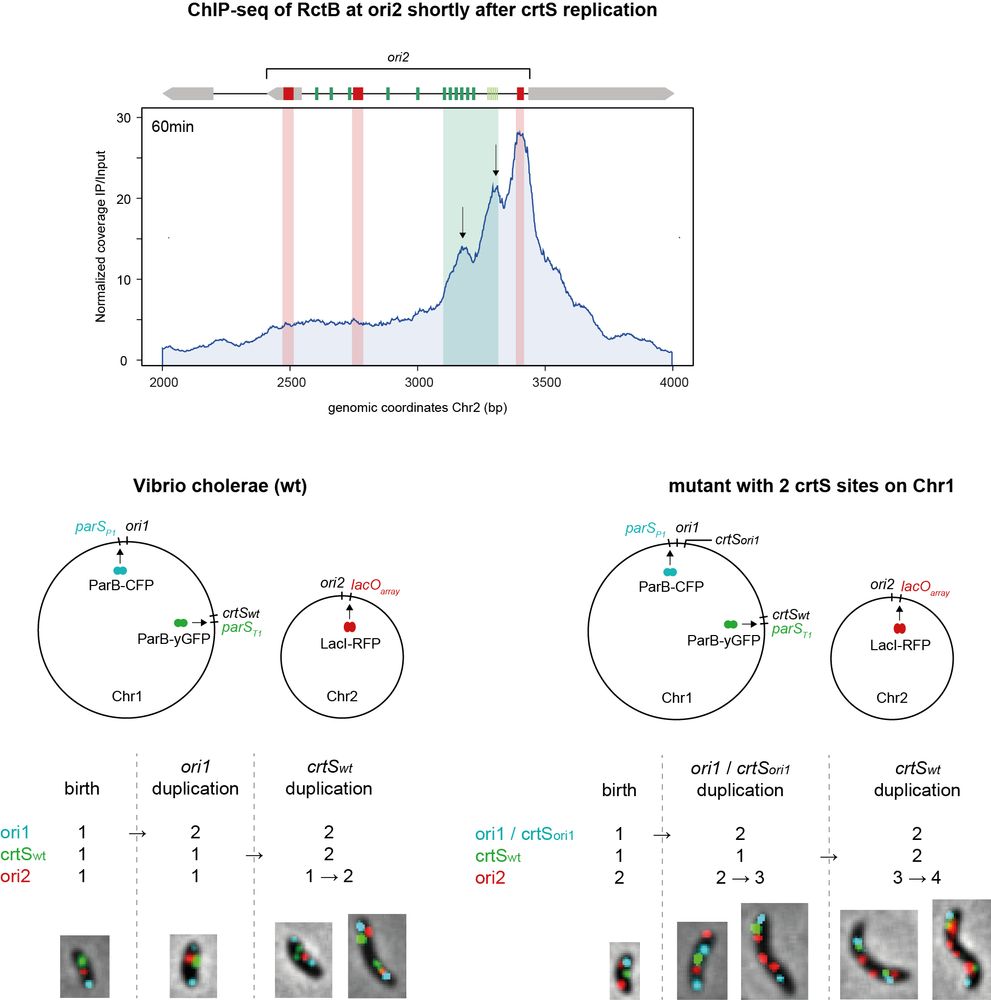
5/ 🗝️ crtS: The key
After crtS is replicated on Chr1, RctB shifts to activation sites (iterons), unlocking Chr2 replication. Each crtS replication event triggers replication of a single ori2. This ensures a precise one-to-one synchronization between Chr1 and Chr2.
After crtS is replicated on Chr1, RctB shifts to activation sites (iterons), unlocking Chr2 replication. Each crtS replication event triggers replication of a single ori2. This ensures a precise one-to-one synchronization between Chr1 and Chr2.
January 9, 2025 at 12:47 PM
5/ 🗝️ crtS: The key
After crtS is replicated on Chr1, RctB shifts to activation sites (iterons), unlocking Chr2 replication. Each crtS replication event triggers replication of a single ori2. This ensures a precise one-to-one synchronization between Chr1 and Chr2.
After crtS is replicated on Chr1, RctB shifts to activation sites (iterons), unlocking Chr2 replication. Each crtS replication event triggers replication of a single ori2. This ensures a precise one-to-one synchronization between Chr1 and Chr2.
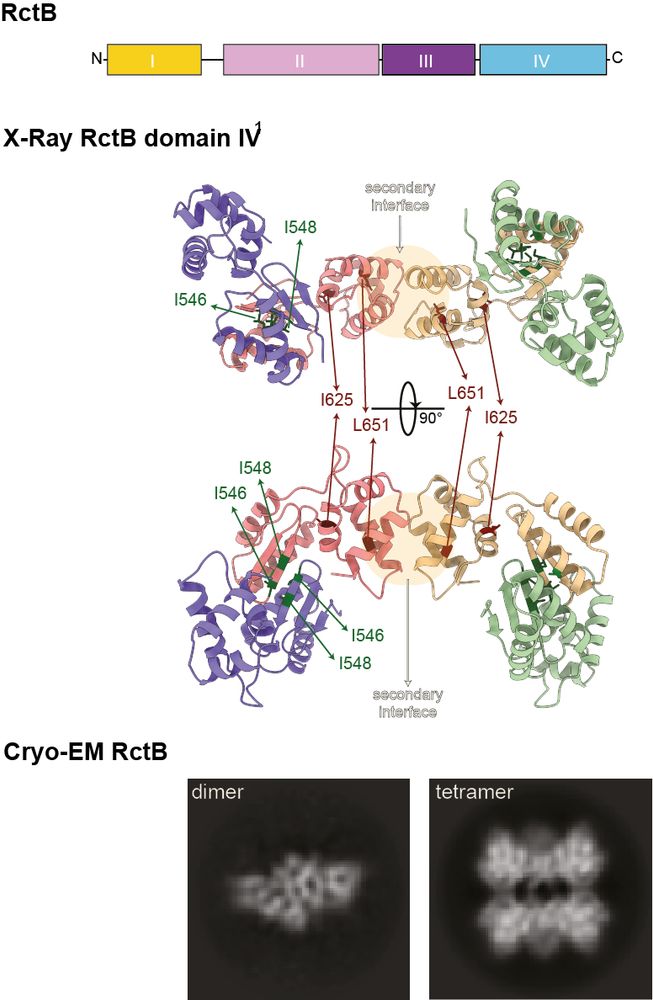
4/ 🔒 RctB: The gatekeeper (part II)
Cryo-EM and X-ray crystallography revealed that RctB oligomerizes through two key interfaces in its domain IV, enabling the formation of higher-order complexes that stabilize the inhibitory DNA loops at ori2.
Cryo-EM and X-ray crystallography revealed that RctB oligomerizes through two key interfaces in its domain IV, enabling the formation of higher-order complexes that stabilize the inhibitory DNA loops at ori2.
January 9, 2025 at 12:46 PM
4/ 🔒 RctB: The gatekeeper (part II)
Cryo-EM and X-ray crystallography revealed that RctB oligomerizes through two key interfaces in its domain IV, enabling the formation of higher-order complexes that stabilize the inhibitory DNA loops at ori2.
Cryo-EM and X-ray crystallography revealed that RctB oligomerizes through two key interfaces in its domain IV, enabling the formation of higher-order complexes that stabilize the inhibitory DNA loops at ori2.
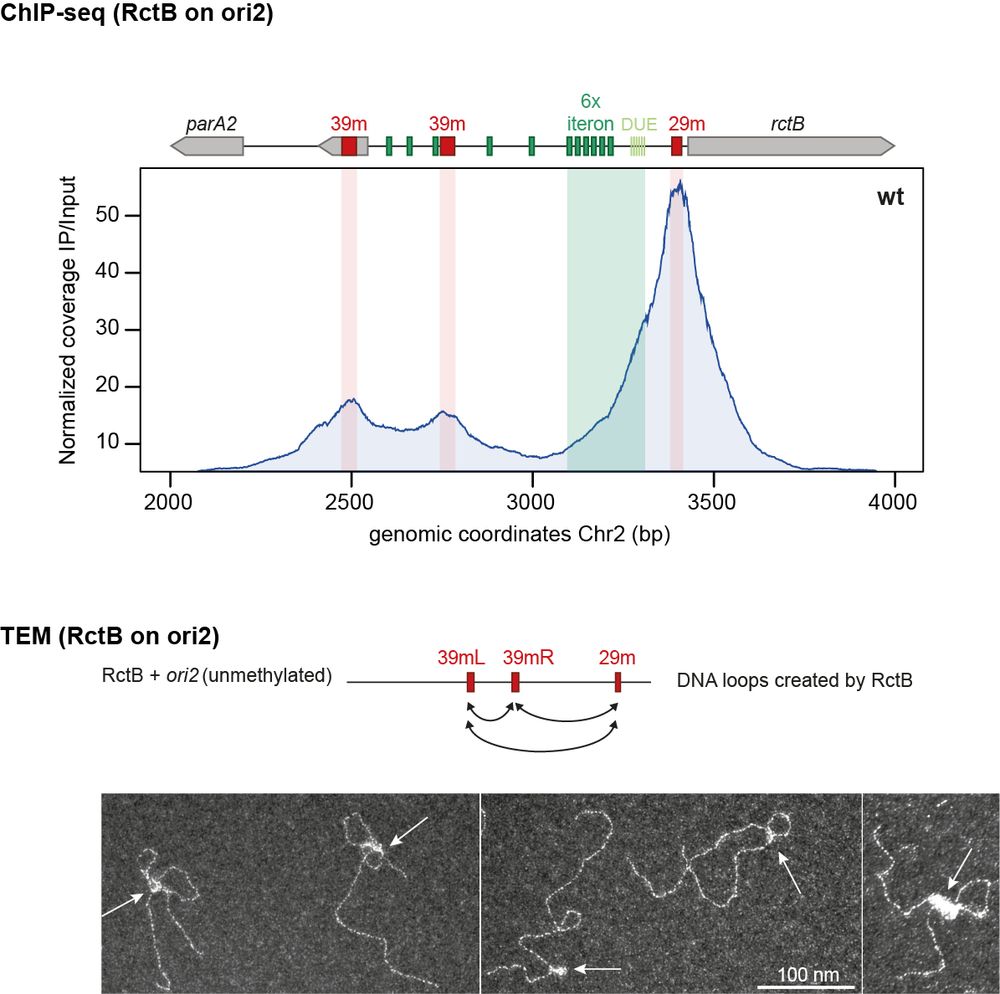
4/ 🔒 RctB: The gatekeeper (part I)
ChIP-seq revealed that RctB binds to inhibitory sites (29/39m) on Chr2’s origin, blocking Chr2 replication for most of the cell cycle. Using TEM, we observed how RctB forms large nucleoprotein complexes at ori2. These complexes bridge 29/39m sites, creating loops.
ChIP-seq revealed that RctB binds to inhibitory sites (29/39m) on Chr2’s origin, blocking Chr2 replication for most of the cell cycle. Using TEM, we observed how RctB forms large nucleoprotein complexes at ori2. These complexes bridge 29/39m sites, creating loops.
January 9, 2025 at 12:45 PM
4/ 🔒 RctB: The gatekeeper (part I)
ChIP-seq revealed that RctB binds to inhibitory sites (29/39m) on Chr2’s origin, blocking Chr2 replication for most of the cell cycle. Using TEM, we observed how RctB forms large nucleoprotein complexes at ori2. These complexes bridge 29/39m sites, creating loops.
ChIP-seq revealed that RctB binds to inhibitory sites (29/39m) on Chr2’s origin, blocking Chr2 replication for most of the cell cycle. Using TEM, we observed how RctB forms large nucleoprotein complexes at ori2. These complexes bridge 29/39m sites, creating loops.
3/ 🧩 The methods
To investigate this mechanism, we mapped the binding of RctB (the initiator for Chr2 replication) across the genome at different points in the cell cycle using ChIP-seq. This was combined with live-cell microscopy, transmission electron microscopy and structural studies.
To investigate this mechanism, we mapped the binding of RctB (the initiator for Chr2 replication) across the genome at different points in the cell cycle using ChIP-seq. This was combined with live-cell microscopy, transmission electron microscopy and structural studies.
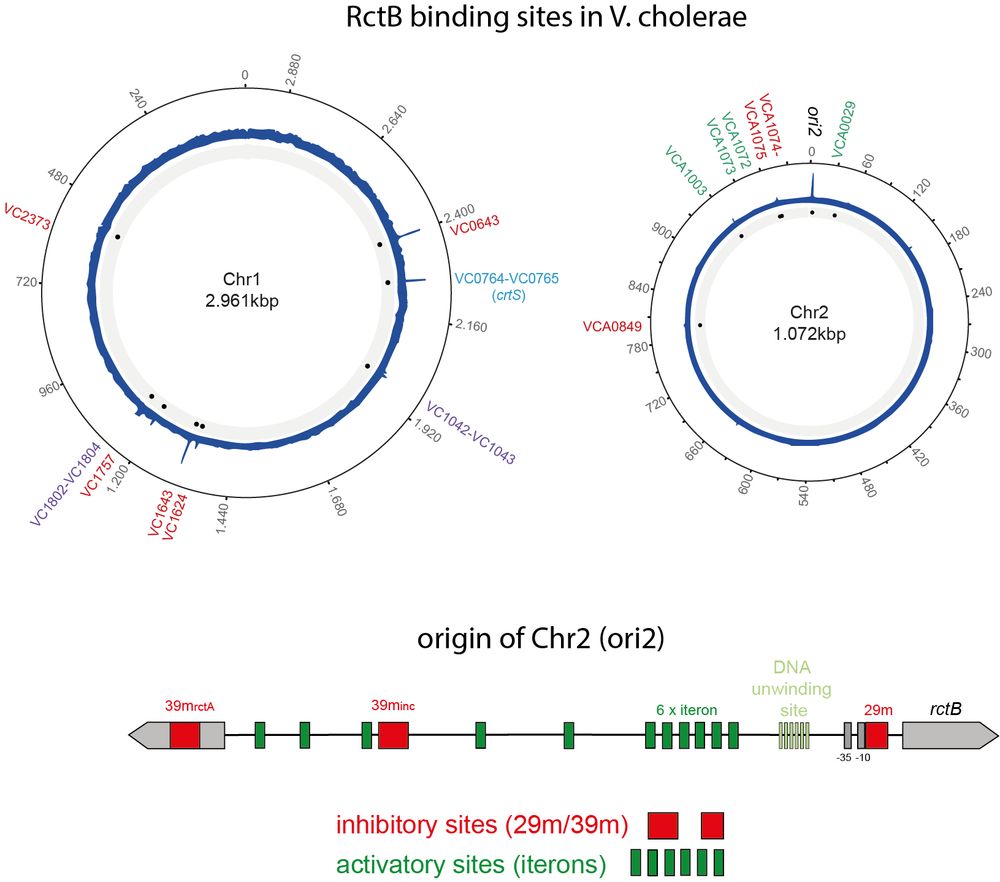
January 9, 2025 at 12:39 PM
3/ 🧩 The methods
To investigate this mechanism, we mapped the binding of RctB (the initiator for Chr2 replication) across the genome at different points in the cell cycle using ChIP-seq. This was combined with live-cell microscopy, transmission electron microscopy and structural studies.
To investigate this mechanism, we mapped the binding of RctB (the initiator for Chr2 replication) across the genome at different points in the cell cycle using ChIP-seq. This was combined with live-cell microscopy, transmission electron microscopy and structural studies.
1/ 🔬 Our work, "Dynamic transitions of initiator binding coordinate the replication of the two chromosomes in Vibrio cholerae", is now published in Nature Communications.
Here's a thread on how we think Chr1 and Chr2 replication is coordinated in Vibrio. 🧵
Link : www.nature.com/articles/s41...
Here's a thread on how we think Chr1 and Chr2 replication is coordinated in Vibrio. 🧵
Link : www.nature.com/articles/s41...

January 9, 2025 at 12:32 PM
1/ 🔬 Our work, "Dynamic transitions of initiator binding coordinate the replication of the two chromosomes in Vibrio cholerae", is now published in Nature Communications.
Here's a thread on how we think Chr1 and Chr2 replication is coordinated in Vibrio. 🧵
Link : www.nature.com/articles/s41...
Here's a thread on how we think Chr1 and Chr2 replication is coordinated in Vibrio. 🧵
Link : www.nature.com/articles/s41...

