@yhgrad.bsky.social @jameshay.bsky.social
End/
@yhgrad.bsky.social @jameshay.bsky.social
End/
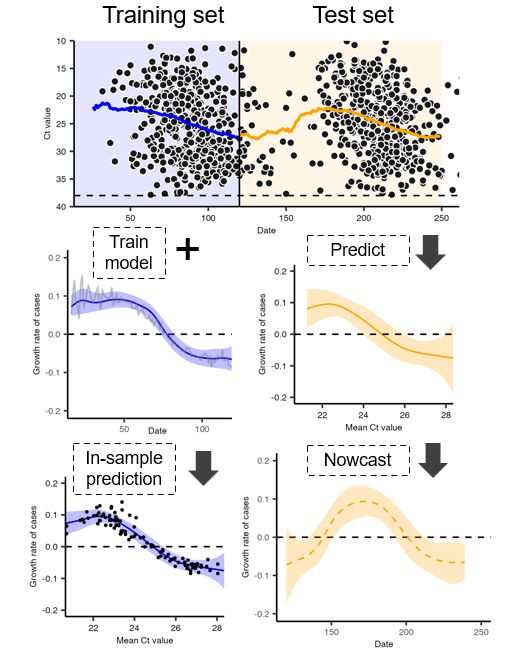
11/
11/
10/
10/
Dropping Ct outliers in each day’s data improves performance, but accounting for population immunity or symptom status does not; all models face clear bias-variance tradeoffs
9/
Dropping Ct outliers in each day’s data improves performance, but accounting for population immunity or symptom status does not; all models face clear bias-variance tradeoffs
9/
8/
8/
7/
7/
6/

6/
5/
5/
4/
4/
3/

3/
2/

2/

